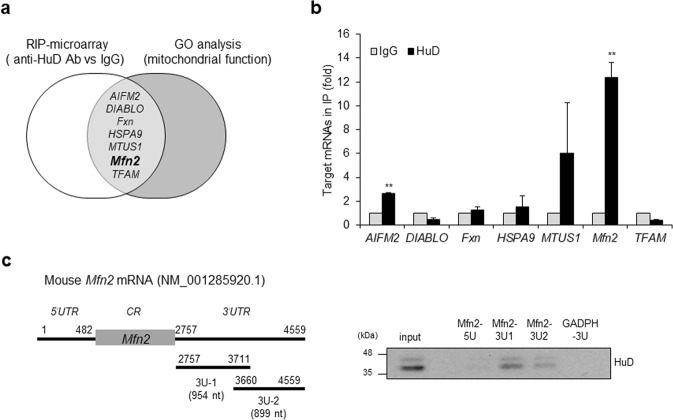
Fig. 4.
Identification of target mRNAs of HuD. a Putative targets of HuD identified by both HuD-RIP analysis and GO term analysis (mitochondrial function). b The interaction between HuD and target mRNAs was confirmed by RIP-qPCR using anti-HuD and control IgG antibodies. Gapdh mRNA was used for normalization. c Left: A schematic of mouse Mfn2 mRNA (NM_001285920). The 5′UTR (5U) and 3′UTR of Mfn2 mRNAs (3U-1 and 3U-2) were transcribed in vitro and biotinylated for use in biotin pull-down analysis. Right: The biotinylated transcripts obtained from Mfn2 mRNA were incubated with βTC6 cell lysates. The interaction between the transcripts and HuD was shown by western blot. Biotinylated Gapdh 3′UTR transcript was used as a negative control. Data represent the mean ± SEM derived from three independent experiments. **p < 0.01