Fig. 2.
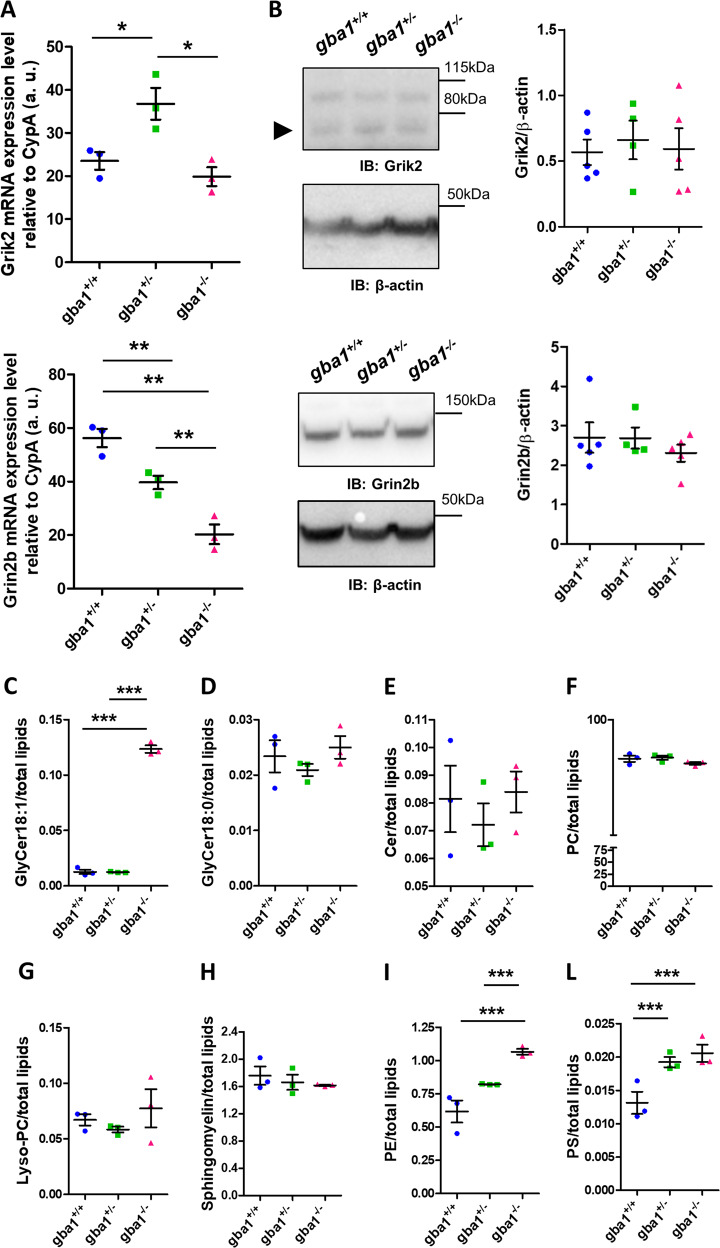
Levels of expression of glutamate receptors and massive accumulation of glycosylceramides are not responsible for increased sensitivity to glutamate in gba1+/− and gba1−/− neurons. a Expression of the glutamate receptors Grik2 and Grin2b in gba1+/+, gba1+/−, and gba1−/− mice brains was measured at the mRNA level by qPCR (shown as scatter plot, mean ± SEM, n = 3 brains per genotype). These data show that Grik2 mRNA was slightly higher in gba1+/− compared with the other genotypes, while Grin2b mRNA levels were decreased in gba1+/− and gba1−/− compared with gba1+/+, showing that the increased glutamate sensitivity is not attributable to increased glutamate receptor expression (One-way Anova, post-hoc Bonferroni, **p < 0.01). b Protein expression of the glutamate receptors Grik2 and Grin2b in gba1+/+, gba1+/−, and gba1−/− mice brains were measured by western blot (shown as scatter plot, mean ± SEM, n = 4–5 brains per genotype). These data show that the expression of these receptors was unchanged at the protein level (One-way Anova, post-hoc Bonferroni). c–m Analysis of lipidomics performed by mass spectrometry on lipids extracted from gba1+/+, gba1+/−, and gba1−/− brains (n = 3 brains per genotype, data represented as scatter plot and mean ± SEM). Glycosylceramide (d18:1) levels were increased in gba1−/− but not in gba1+/+ and gba1+/− brains (One-way Anova, post-hoc Bonferroni, ***p < 0.001) (c), suggesting that the enzyme activity produced by a single copy of GBA1 gene is sufficient to avoid substrate accumulation. Curiously, glycosylceramide (d18:0) levels were unchanged (d), as well as Cer, PC, lyso-PC, SM (e–h), while PE and PS are significantly changed in the disease models compared with wild-type brains (i–l)