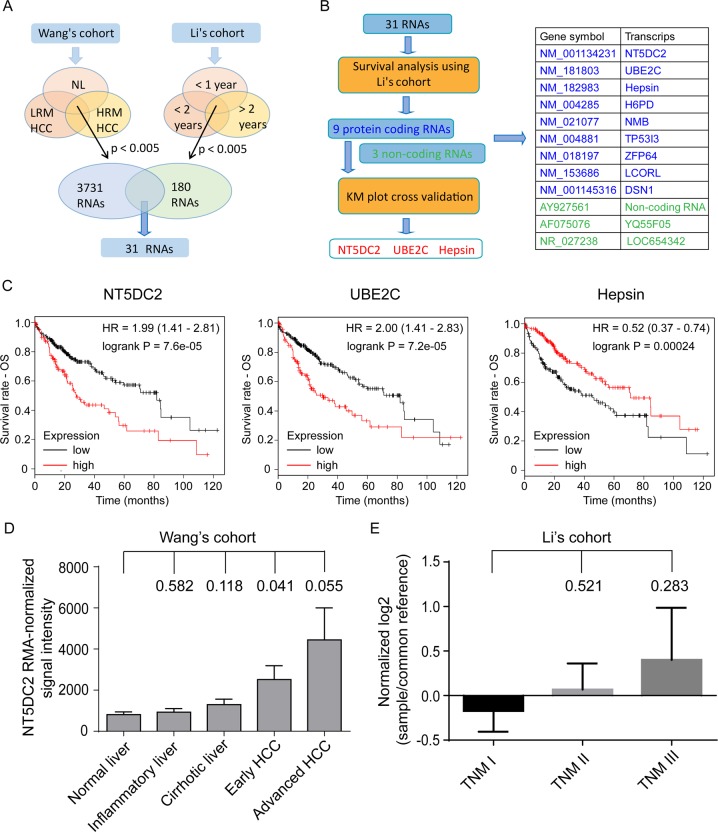
Fig. 1. Identification of NT5DC2 as a potential target for treating HCC.
a Schematic graph showing the identification of differentially expressed RNAs after analyzing two GEO databases (GSE54238 for Wang’s cohort, GSE40144 for Li’s cohort). b Flowchart showing the identification of NT5DC2, UBE2C and hepsin as candidate genes. Twelve genes were found significantly associated with OS and DFS in Li’s cohort. Further cross-validation of patient OS using K–M plot identified NT5DC2, UBE2C and hepsin as candidate genes. c Candidate genes significantly associated with patient’s OS in K–M plot database (http://www.kmplot.com/). d NT5DC2 expression in normal liver, inflammatory liver, liver with cirrhosis, early HCC and advanced HCC in Wang’s cohort. Student’s t-tests were performed to analyze the differences between each group and normal liver. P values are labeled above the column for each group. e NT5DC2 expression in TNM stage I patients, stage II patients and stage III patients in Li’s cohort. Student’s t-tests were performed to analyze the differences between each group and TNM stage I patients. P values are labeled above the column for each group.