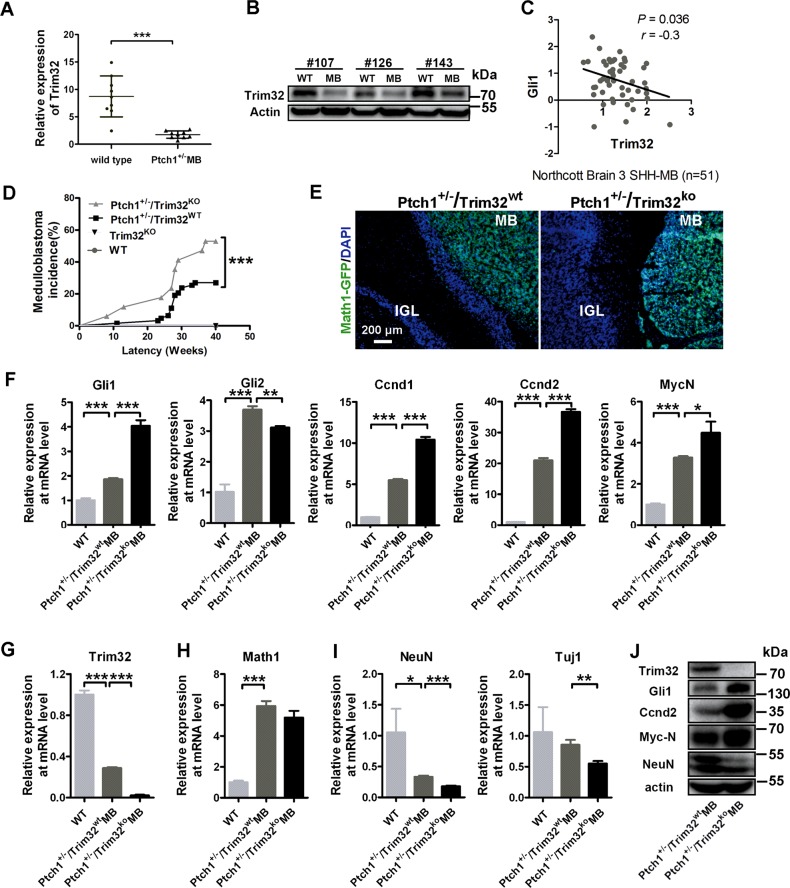
Fig. 6.
Trim32 knockout increases the incidence of MB in Ptch1+/− mice. Quantitative PCR mRNA (a; mean ± SD; n = 9) and protein levels (b; n = 3) of Trim32 in mouse medulloblastomas (MB) from Ptch1+/− mice and normal cerebella. c RNA expression level of Trim32 negatively correlated with Gli1 (n = 51) in human SHH MB samples. Data are analyzed from Oncomine database. d Kaplan–Meier analysis of MB incidence in 63 Ptch1+/−/Trim32wt mice (gray line) versus 17 Ptch1+/−/Trim32KO mice (black line). Ptch1+/−/Trim32wt mice and Ptch1+/−/Trim32KO mice, obtained by interbreeding for at least three generations the progeny of Ptch1 heterozygous and Trim32 knock-out mice, were then monitored for the onset of medulloblastoma; (triple asterisks indicate P < 0.001, Logrank test). e Expression of the Math1-GFP protein (GFP, green) in the Math1-GFP mouse medulloblastomas and adjacent cerebellar cortices sections from Ptch1+/−/Trim32wt mice and Ptch1+/−/Trim32KO mice. Nuclei were counterstained with DAPI (blue). The scale bar represents 200 µm. MB medulloblastoma, IGL internal granule layer. RT-qPCR analysis of SHH target genes (f), Trim32 (g), the granule neuronal progenitor marker Maht1 (h), and the differentiated granule cell markers including Tuj1 and NeuN (i) in adult mouse cerebellums and medulloblastomas from Ptch1+/−/Trim32wt mice and Ptch1+/−/Trim32KO mice. Data are expressed as means ± SD (n = 3). An asterisk indicates P < 0.05, double asterisks indicate P < 0.01, and triple asterisks indicate P < 0.001. j Immunoblotting analysis of Trim32, Gli1, Ccnd2, MycN, and NeuN in MB from Ptch1+/−/Trim32wt mice and Ptch1+/−/Trim32KO mice