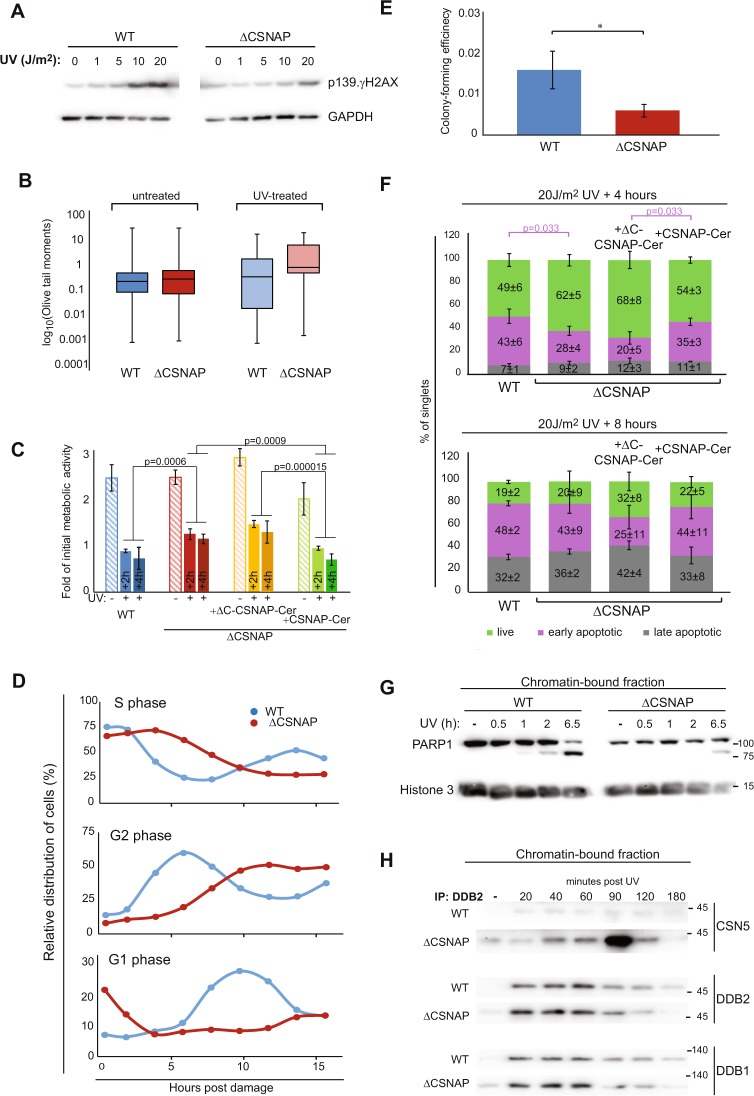
Fig. 4.
Recovery following exposure to UV irradiation is affected by the absence of CSNAP. a DNA damage response is attenuated in ΔCSNAP cells. Analysis of dose-dependent induction of γH2AX Ser139 phosphorylation. Western blots were performed 1.5 h post damage. Representative results out of five experiments indicating reduced γH2AX Ser139 phosphorylation in ΔCSNAP cells. b Unlike WT cells, damaged DNA accumulates in ΔCSNAP cells following UV exposure. The genotoxic effect of UV was measured using alkaline comet assay. DNA damage, expressed as Olive tail moments were calculated and presented as a box-whisker plot. Within each biological replicate, the proportion of Olive tail moments larger than 3 was calculated. The proportions in ΔCSNAP and WT samples were compared using a two-way ANOVA, accounting for group and batch (UV treated samples p = 0.00142). Data are shown before and after UV treatment (representative plot of 6 replicates, each). c Comparison of metabolic activity of WT and ΔCSNAP cells before and after UV irradiation. The plot shows metabolic activities of WT and ΔCSNAP cells, before and after UV induced DNA damage, 26 and 28 h (2 and 4 h post UV, respectively) after seeding, calculated as a fold of initial activity for each cell line. Metabolic activity of both WT and ΔCSNAP cells is reduced in response to DNA damage but a difference in the decrease in activity is detected, a pronounced degree of metabolic reduction is observed for WT cells or ΔCSNAP cells expressing full length CSNAP-Cerulean. The graph represents the averages of three independent experiments, with standard errors. Significance was calculated using a two-way ANOVA test accounting for treatment, time, cell type, and batch (effect of treatment: p < 2e−16, effect of time: p = 0.07905, effect of cell type: p = 1.27e−08), another ANOVA was run only on the treated samples, followed by a Tukey's post hoc test for cell type. d UV-exposed ΔCSNAP cells stay longer in S and G2 phases. Comparison of the relative distribution of cell populations in different phases of the cell cycle, as calculated from flow cytometry histograms of double thymidine-synchronized cells following exposure to UV irradiation (5 J/m2). UV-induced DNA damage causes longer cell cycle phases in cells lacking CSNAP. The graph shows a representative experiment out of three. e Cells lacking CSNAP exhibit a compromised recovery after exposure to high-dose UV. WT and ΔCSNAP cells were exposed to UV irradiation, prior to incubation in culturing conditions for 8 days. Colonies were stained and counted. ΔCSNAP cells exhibit ~2.7-fold less colony-forming potential following UV damage, in comparison with WT cells. The graph represents average results from seven biological replicates with standard errors. Significance was calculated using paired Student’s t-test (p = 0.035). f The early apoptotic response is delayed in ΔCSNAP cells, following UV damage. The bar charts represent the average percentage of live, early, and late apoptotic cells detected by three independent flow cytometry experiments for each time points ± standard errors. A significant difference is seen in the percentage of early apoptotic cell populations between WT and ΔCSNAP cells after 4 h of UV exposure, calculated using two-way ANOVA, accounting for treatment and batch (effect of treatment in early apoptosis p = 0.00506) followed by a Tukey’s post hoc test. Overexpression of CSNAP-Cerulean in cells lacking CSNAP rescues the late-onset apoptosis, but not when its C-terminal interacting domain is missing. Eight hours after UV irradiation the distribution of apoptotic cells is similar in both WT and ΔCSNAP cells. g PARP1 cleavage is delayed in cells lacking CSNAP. Chromatin-bound fractions were monitored by western blot for caspase-mediated PARP1 cleavage, a marker for commitment to apoptosis. h CSNΔCSNAP exhibits increased affinity toward DDB2, in comparison with the CSN complex. WT and ΔCSNAP cells were exposed to UV irradiation, and DDB2 was immunoprecipitated from the chromatin-bound fraction at different time points post UV damage. Western blot analyses show tighter CSN-CRL binding when CSNAP is absent. Representative blot out of four repeats