Figure 4.
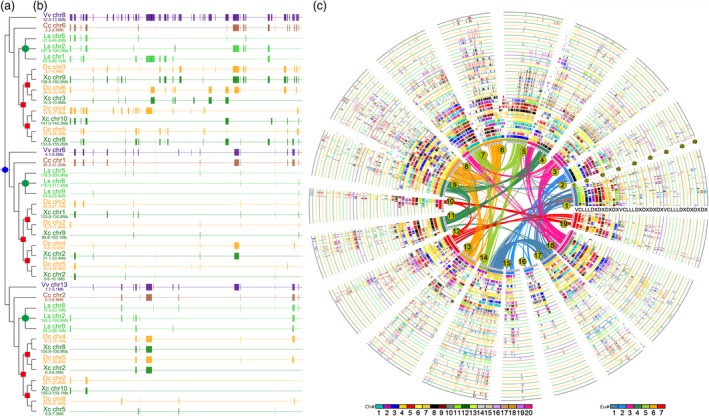
Local and global alignment of genomes. (a) A gene phylogenetic tree was constructed using colinear genes among coriander, carrot, lettuce, coffee and grape. Hexagons indicate eudicot‐common hexaploidy (ECH, blue), Asteraceae‐common hexaploidy (ACH, green) and the Apiaceae palaeo‐tetraploidization events, A‐beta and A‐alpha (red). (b) Local alignment of genes among coriander (Xc), carrot (Dc), lettuce (Ls), coffee (Cc) and grape (Vv). Using as reference a homoeologous series of grape segments produced by the ECH, we displayed the alignment of a region from 12.3 to 13.5 Mb on grape chromosome 8, 4.1 to 5.5 Mb on chromosome 6 and 1.7 to 3.1 Mb on chromosome 13, along with corresponding regions from other genomes. Chromosome numbers follow the names of plants, and locations on chromosomes are below. Gene (rectangle) positions correspond to those of colinear grape genes. (c) Global alignment of homologous regions in coriander (X), carrot (D), lettuce (L) and coffee (C) genomes with grape (V) as a reference. Genomic paralogy, orthology and outparalogy information within and among genomes are displayed in 39 circles, each corresponding to an extant gene. The curved lines within the inner circle are formed by 19 grape chromosomes colour‐coded to correspond to the 7 ancestral chromosomes before the major eudicot‐common hexaploidy (ECH). The short lines forming the innermost grape chromosome circles represent predicted genes, which have 2 sets of paralogous regions, forming another two circles. Each of the three sets of grape paralogous chromosomal regions has one ortholog in coffee, 3 in lettuce and 4 in coriander and carrot. The resulting 39 circles were marked according to species by a capital letter. Each circle has an underline coloured as to its source plant, corresponding to the colour scheme, and each circle is formed by short vertical lines that denote homologous genes, coloured as to chromosome number in their respective source plant as shown in the inset colour scheme.
