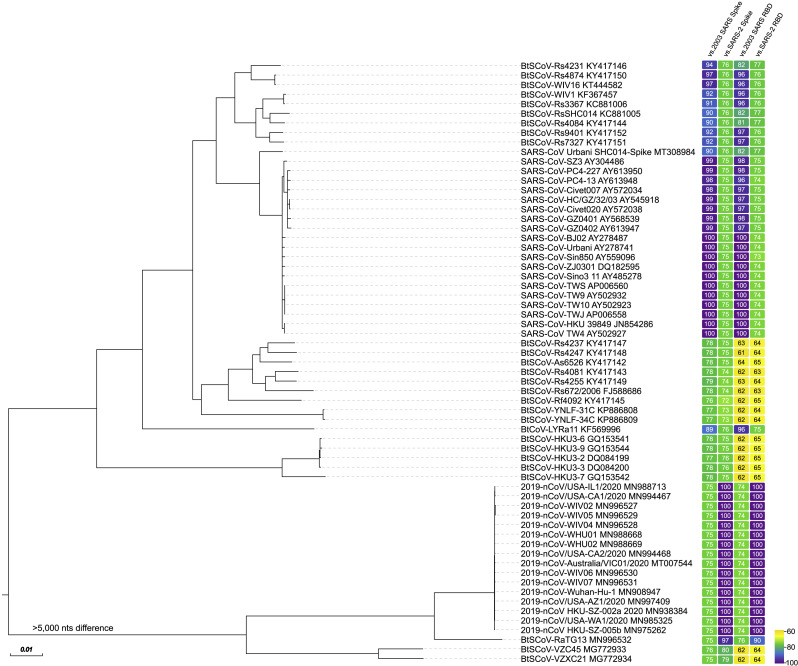
Figure 1.
Genome Phylogeny and Spike and Receptor-Binding Domain Identity of Representative Group 2b Betacoronaviruses
The genome, spike, and RBD sequences of selected group 2b betacoronaviruses were aligned and phylogenetically compared. Sequences were aligned using free end gaps with the Blosum62 cost matrix in Geneious Prime. The phylogenetic tree was constructed from the multiple genome sequence alignment using the neighbor-joining method based on 100 replicates, also in Geneious Prime. The GenBank accession number follows each sequence name. Spike and RBD amino acid sequence identities from their respective alignments are represented by color-coded boxes to the right of each tree position, with colors ranging from yellow (~60% similarity) to purple (~100% similarity), shown in the scale in the lower right. Identities are represented as versus 2003 SARS-CoV spike (listed in figure as “2003 SARS Spike”), versus SARS-CoV-2 spike (“SARS-2 Spike”), versus 2003 SARS-CoV RBD (“2003 SARS RBD”), and versus SARS-CoV-2 RBD (“SARS-2 RBD”). The phylograms and alignments were exported from Geneious and then rendered for publication using EvolView (www.evolgenius.info) and Adobe Illustrator CC 2020.