Figure 1.
Genome Organization in OIS and RS Based on Distinct Chromatin Compartment Interactions
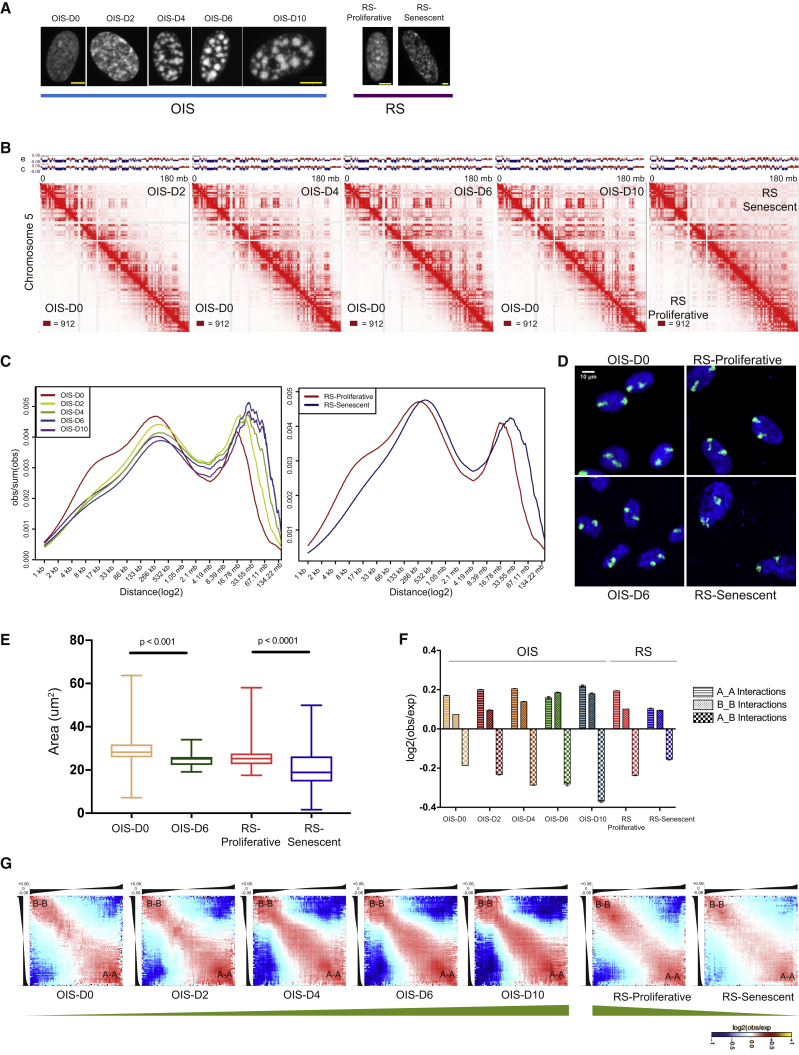
(A) Schematic representation (DAPI staining) of the WI-38 hTERT/GFP-RAF1-ER cell-specific OIS and WI-38 primary cell-dependent RS systems. Only OIS cells show SAHF bodies (D4 onward). Scale bar, 5 μm.
(B) Normalized Hi-C contact maps for chromosome 5 at 500 kb resolution. The bottom left of individual Hi-C plots represents control cells (D0 or RS-Proliferative), and the top-right part displays senescence conditions. The respective eigenvectors are above the Hi-C plot. c, controls; e, senescent conditions. The maximum intensity for each panel is indicated in the bottom-left corner.
(C) Contact probability in logarithmic bins. Lines: mean values from biological replicates.
(D) Representative images of chromosome territories from OIS-D0/OIS-D6 and RS-Proliferative/senescent cells mapped via whole-chromosome paint assay. Chromosome territories were delineated with Cy5 (green)-labeled whole-chromosome painting probes, and the nuclei were counterstained with DAPI (blue). Bar, 10 μm.
(E) The distributions of areas of chromosome territories are shown as boxplots. Statistical significance was calculated using the Mann-Whitney test.
(F) Intra- and inter-compartment contact enrichment from OIS and RS samples. Data represented as bar plots show the mean ± SD.
(G) Average contact enrichment between pairs of 250 kb loci arranged by their eigenvalue (shown on top). The green bar at the bottom depicts the trend in compartmentalization. See also Figure S1.