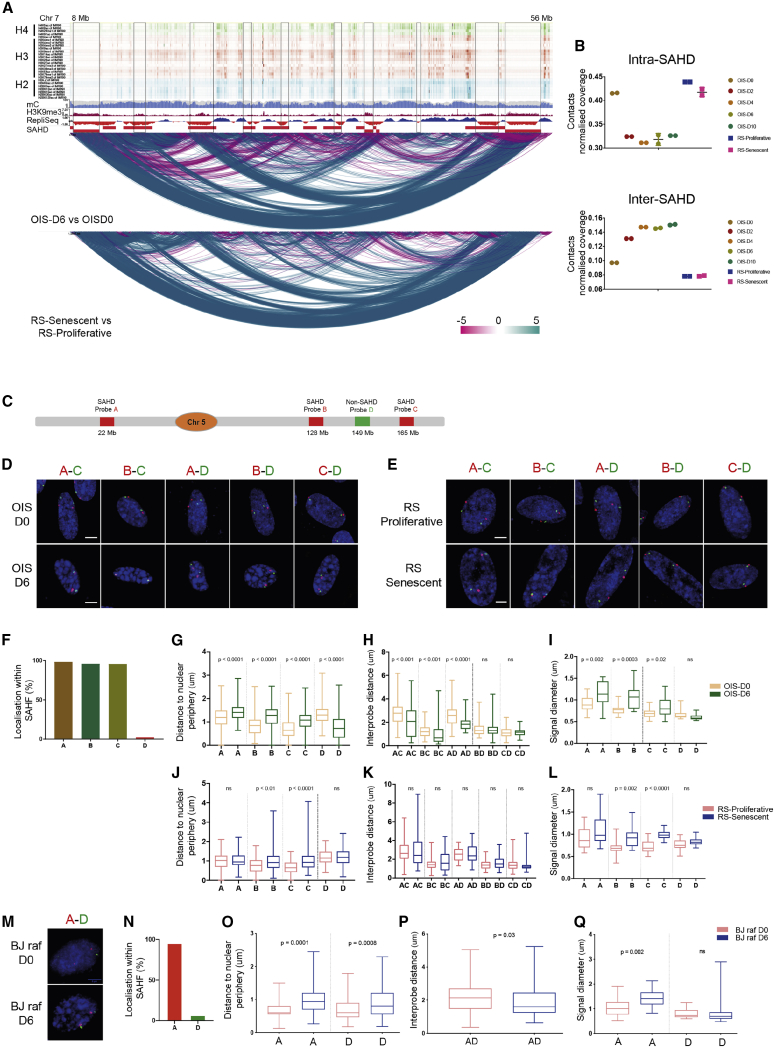
Figure 2.
Differential Intra- versus Inter-SAHD Interactions Constitute SAHF
(A) Genome browser shot of differential interactions. Top: histone modification tracks of H4 (K5ac, K8ac, K20me1, K91ac), H3 (K4me1, K4me2, K4me3, K4ac, K9me1, K9ac, K14ac, K18ac, K23ac, K27me2, K27ac, K36me3, K56ac, K79me1, K79me2), and H2 (A.Z, BK5ac, AK9ac, BK12ac, BK15ac, Bk20ac, BK120ac) modifications from IMR90 cells. Middle: H3K9me3, mC, and Repli-seq tracks from IMR90 cells. Bottom: differential interaction (from diffHiC) tracks from OIS-D0 versus OIS-D6 and RS-Proliferative versus RS-Senescent at 100 kb bins. Red rectangles highlight SAHDs. The color scale bar represents statistically significant differential scores in 100 kb bins. The blue arcs are gains in interaction while purple arcs mean losses.
(B) Quantification of contacts within and between SAHDs in OIS and RS. Data are represented as a scatter dot plot showing the mean ± SD.
(C) Schematic representation of the location of FISH probes on chromosome 5.
(D) Representative 3D-DNA FISH images (z-slice) from OIS-D0 and OIS-D6 samples with indicated probes. Scale bar, 5 μm.
(E) Representative 3D-DNA FISH images (z-slice) from RS-Proliferative and RS-Senescent samples with indicated probes. Scale bar, 5 μm.
(F) Percentage of the respective FISH probes localized within SAHFs in individual nuclei in OIS-D6 samples.
(G) Boxplot quantification of the SAHD (A, B, and C) and non-SAHD (D) probe distance from the nuclear periphery.
(H) Boxplot of inter-probe distances in OIS-D0 and OIS-D6.
(I) Quantification of the changes in the diameter of the individual probes in OIS-D0 and D6.
(J) Quantification showing the distances of the probe from the nuclear periphery in RS-Proliferative and RS-Senescent.
(K) Boxplot of inter-probe distances in RS-Proliferative and RS-Senescent.
(L) Quantification of the changes in the diameter of the individual probes in RS-Proliferative and RS-Senescent.
(M) Representative 3D-DNA FISH images (z-slice) from BJ raf D0 and BJ raf D6 samples with A–D probes. Scale bar, 5 μm.
(N) Percentage of the respective FISH probes, localized within SAHFs in individual nuclei, in BJ raf D6 samples.
(O) Quantification showing the distance of the SAHD (A) and non-SAHD (D) probes from the nuclear periphery. Data are represented as boxplots.
(P) Boxplot of inter-probe distances in BJ raf D0 and BJ raf D6.
(Q) Quantification of the changes in the diameter of the individual probes in BJ raf D0 and BJ raf D6. Statistical significance in (G)–(L) and (O)–(Q) was calculated using the Mann-Whitney test.