Figure 3.
Detachment from the Lamina Partially Explains 3D Genome Reorganization in OIS
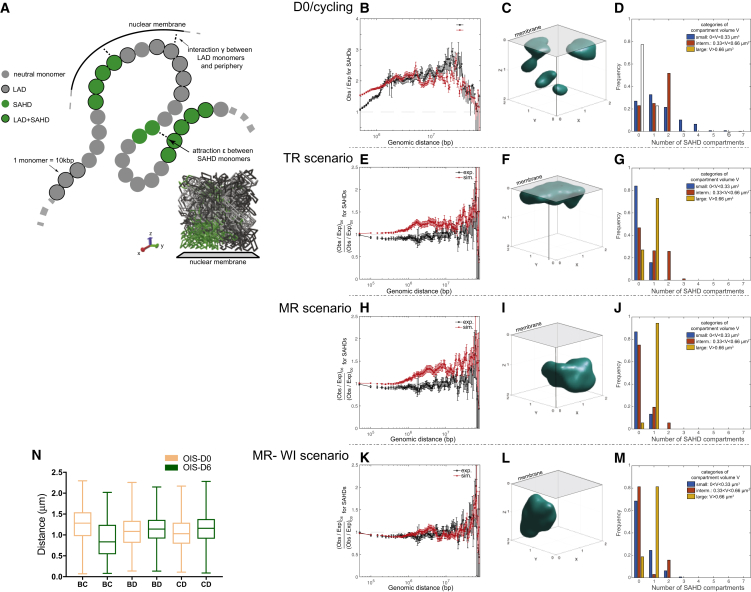
(A) A two-parameter polymer model of heterochromatin organization and positioning. Each 10 kbp monomer is characterized by an epigenetic and lamina-interacting state. SAHD monomers exhibit pairwise contact attraction and cLAD-like loci may interact preferentially with the nuclear membrane. An example of configuration evolving in a cubic box (one of its face representing the membrane) is given at the bottom-right corner of the figure.
(B–D) The inference of a polymer model for cycling/OISD0 cells. (B) Ratio between the average contact probability between SAHD regions and the total average contact probability between any pairs of loci (expected probability), as a function of the genomic distance. (C) Example of SAHD compartment. (D) Distributions of the number of small, intermediate, and large SAHF-like SAHD compartments per simulated configuration.
(E–G) Display the predictions for the time-release (TR) scenario. (E) Ratio between the observed versus expected ratio for SAHD regions at OIS-D6 and at OIS-D0. (F) Example of SAHD compartment. (G) Distributions of the number of small, intermediate, and large SAHF-like SAHD compartments per simulated configuration.
(H)–(J) Predictions for the membrane-release (MR) scenario. (H) Ratio between the observed versus expected ratio for SAHD regions at OIS-D6 and at D0. (I) Example of SAHD compartment. (J) Distributions of the number of small, intermediate, and large SAHF-like SAHD compartments per simulated configuration.
(K–M) Predictions for the membrane-release with weakening of SAHD attraction (MR-WI) scenario. (K) Ratio between the observed versus expected ratio for SAHD regions at OIS-D6 and at OIS-D0. (L) Example of SAHD compartment. (M) Distributions of the number of small, intermediate, and large SAHF-like SAHD compartments per simulated configuration.
(N) Distance distribution between B, C, and D probes from the polymer model.
See also Figure S3.