Figure 5.
DNMT1 Is Associated with OIS-Dependent 3D Genome Rewiring
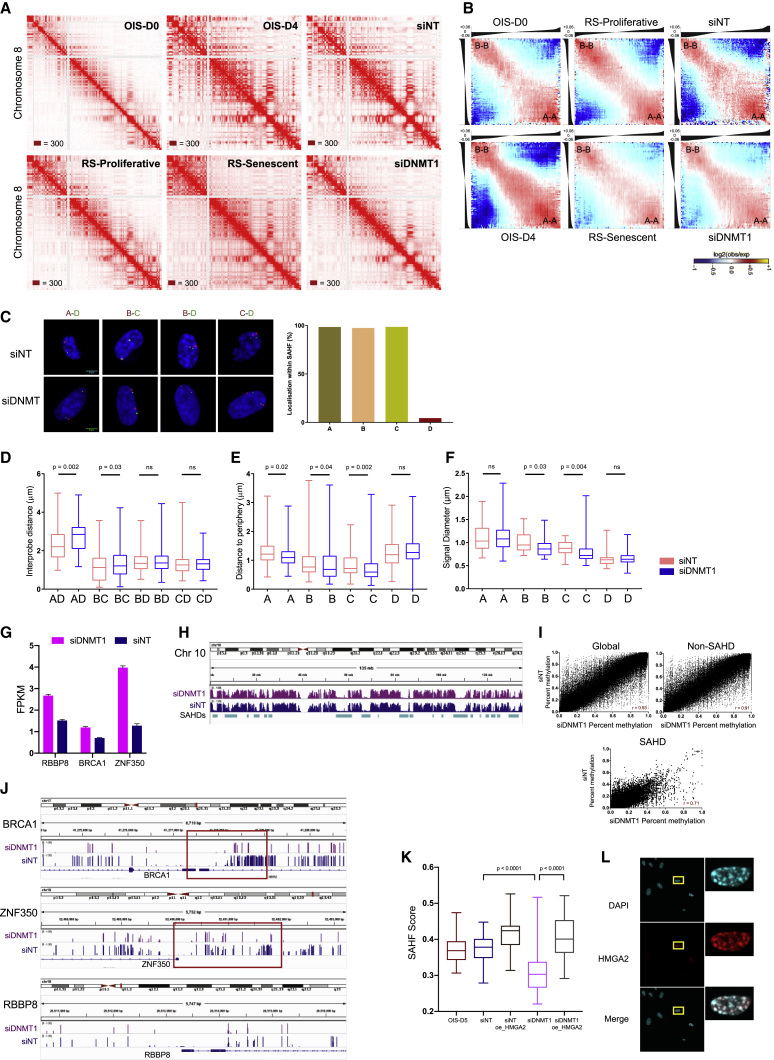
(A) Normalized Hi-C contact maps shown for chromosome 5 at 500 kb resolution. The maximum intensity for each data point is indicated in the bottom-left corner.
(B) Average contact enrichment between pairs of 250 kb loci arranged by their eigenvalue (shown on top).
(C) The left panel is representative 3D-DNA FISH images (z-slice) from NT and DNMT1-depleted samples with A-C, B-C, B-D, and C-D probes. Scale bar, 5 μm. The right panel displays the percentage of the respective FISH probes, localized within SAHFs in individual nuclei, in NT samples.
(D) Boxplot of inter-probe distances in NT and DNMT1-depleted samples.
(E) Quantification showing the distance of the SAHD (A, B, and C) and non-SAHD (D) probe from the nuclear periphery. Data are represented as boxplots.
(F) Quantification of the changes in the diameter of the individual probes in NT- and DNMT1-depleted samples. Statistical significance in (D)–(F) was calculated using the Mann-Whitney test.
(G) BRCA1, ZFP350, and RBBP8 expression represented as the mean ± SD of two biological replicates of RNA sequencing (RNA-seq) experiments for NT and DNMT1-depleted samples.
(H) IGV (Integrative Genomics Viewer) snapshot of chromosome 10 with WGBS tracks from siNT- and siDNMT1-treated OIS samples.
(I) Scatterplot comparing the mC levels (percentage of methylation) in siNT and siDNMT1 samples. The correlation of the individual datasets is indicated on the bottom-right panel.
(J) IGV snapshot of the -BRCA-1, ZNF350 and RBBP8 genes, along with WGBS tracks from NT- and DNMT1-depleted OIS cells. The red box highlights the differentially methylated promoter region.
(K) Quantification of the changes in the SAHF score (details in STAR Methods). Statistical significance was calculated using the Mann-Whitney test.
(L) An immunofluorescence experiment on OISD5 cells in DNMT1-deleted and HMGA2 overexpression condition using an antibody against HMGA2. The zoomed cell displays a single nucleus with SAHF bodies and HMGA2 overexpression.
See also Figure S5.