Figure 6.
Gene Expression Changes Associated with SAHF Formation
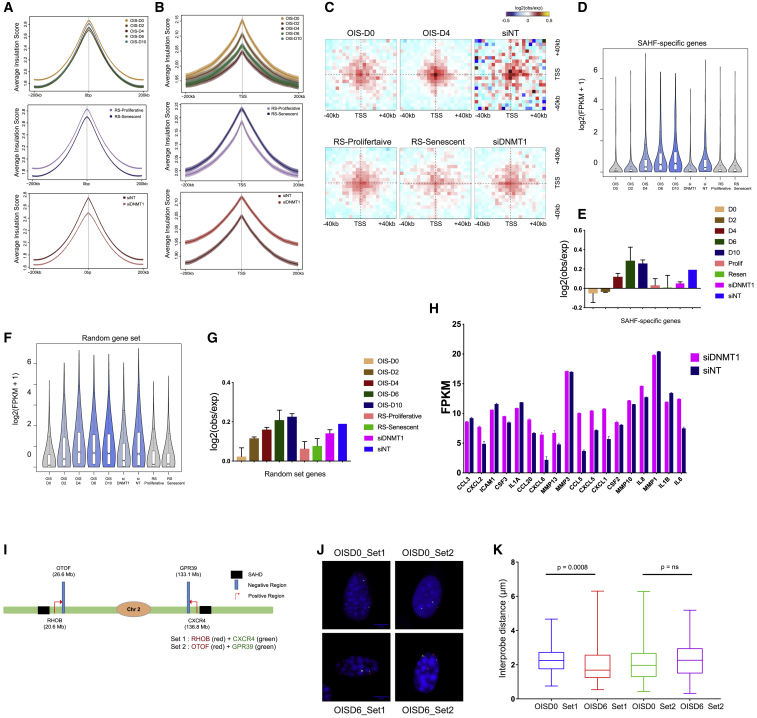
(A) Average insulation score over the 200 kb region around TAD boundaries in OIS, RS, and siRNA-treated OIS cells. Lines show mean values, while dark and light shaded ribbons represent SD and 95% confidence interval (CI), respectively.
(B) Average insulation score centered on gene promoters in OIS, RS, and siRNA-treated OIS cells. Lines show mean values; dark and light shaded ribbons represent SD and 95% CI, respectively.
(C) Long-range (2- to 10-Mb) inter-TAD aggregate Hi-C contact maps around pairs of active transcription start sites (TSS) in OIS-D0, OIS-D2, OIS-D4, RS-Proliferative, RS-Senescent, NT-depleted, and DNMT1-depleted samples within ±40 kb.
(D) Quantification of gene expression of the SAHF-specific genes following the STAR Methods. The data are displayed as boxplot of log 2(FPKM +1) values.
(E) Boxplot quantification (showing the mean ± SD) of the contact strength between pairs of TSS from SAHF-specific genes represented in (D).
(F) Quantification of gene expression of the random ADJ control genes (random gene set). The data are shown as a boxplot of log 2(FPKM +1) values.
(G) Boxplot quantification (showing the mean ± SD) of the contact strength between pairs of TSS from random gene-set category.
(H) SASP gene expression represented as the mean ± SD of two biological replicates of RNA-seq experiments for NT and DNMT1-depleted OIS cells.
(I) Schematic representation of the location of FISH probes on chromosome 2.
(J) Representative 3D-DNA FISH images (z-slice) from OIS-D0 and OIS-D6 samples with set 1 (RHOB and CXCR4) and set 2 (OTOF and GPR39) probes. Scale bar, 5 μm.
(K) Boxplot quantification showing the distance of the within each probe set. Statistical significance was calculated using the Mann-Whitney test.
See also Figure S6.