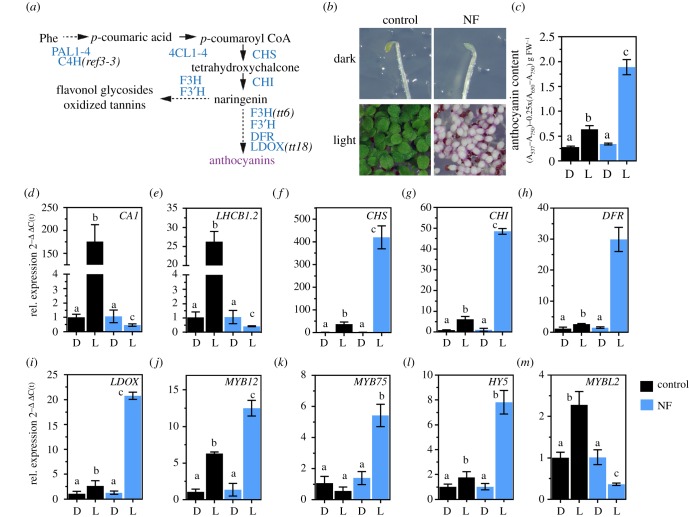
Figure 1.
Induction of flavonoid/anthocyanin biosynthesis (FAB) upon chloroplast dysfunction. (a) Schematic overview of the FAB pathway leading to the formation of polyphenolic compounds. PAL1-4, PHENYLALANINE AMMONIA LYASE; C4H, CINNAMIC ACID 4-HYDROXYLASE; 4CL1-4, 4-COUMAROYL COA LIGASE 1-4; CHS, CHALCONE SYNTHASE; CHI, CHALCONE ISOMERASE; F3H, FLAVANONE 3-HYDROXYLASE; F3'H, FLAVONOID 3′-HYDROXYLASE; DFR, DIHYDROFLAVONOL-4-REDUCTASE; LDOX, LEUCOANTHOCYANIDIN DIOXYGENASE. (b) Representative photograph of etiolated (dark) and continuous light-grown Arabidopsis thaliana Col-0 seedlings germinated in the absence (control) or presence of norflurazon (NF). (c) Quantification of anthocyanins in seedlings shown in (b). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). FW, fresh weight. Black (control) and blue (NF). (d–m) qRT-PCR analysis of gene expression for genes encoding PHANGs (CARBONIC ANHYDRASE1 and LIGHT HARVESTING COMPLEX B1.2), enzymes involved in FAB and transcription factors (MYB12, MYB75, HY5, MYBL2). Seedlings were grown as described in (b). D, dark; L, light. Gene expression was calculated relative to the WT etiolated in the absence of NF and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05).