Figure 5. Conceptual classification of SNVs based on their functional impact and selection characteristics, and additive effects model:
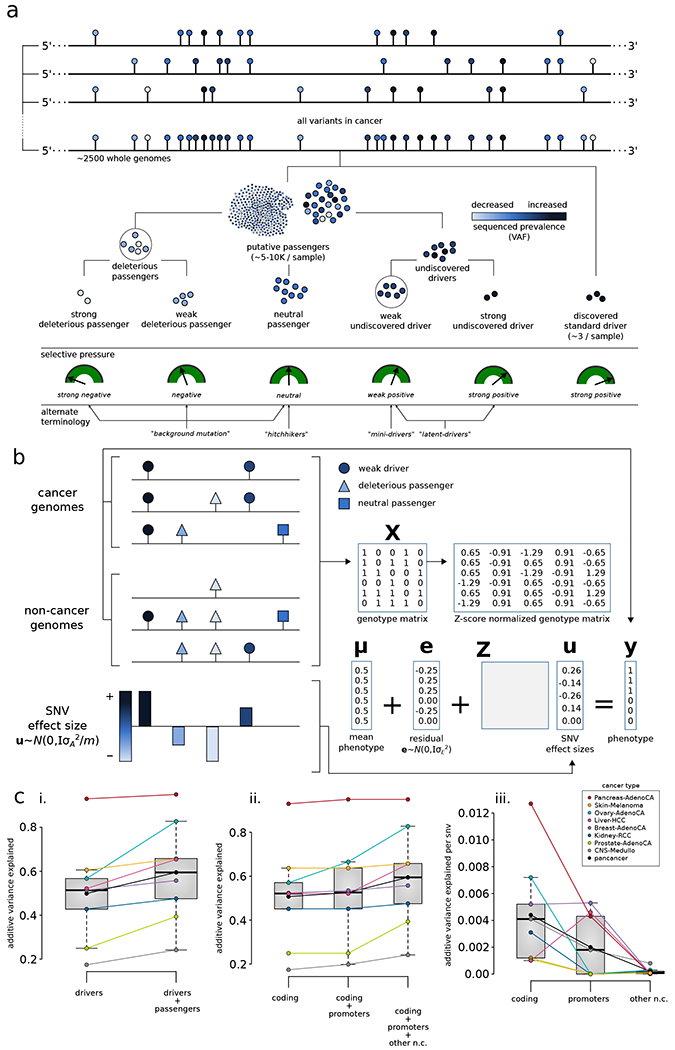
a) In addition to canonical drivers, deleterious passengers (weak and strong) and mini drivers (weak and strong) represent additional categories of cancer mutations in the extended model. b) Additive effects model for putative passengers: The combined effect of many nominal passengers is modeled linearly and predicts whether a genotype arises from an observed cancer sample or from a null (neutral) model. c) Predictive power of known drivers and putative passengers using the additive effects model: (i) compares the maximum possible variance that can be explained using known drivers; (ii) further splits the variance into contributions from coding, non-coding, and promoter variants; (iii) presents normalized additive variance explained exclusively by putative passengers in coding regions, by promoters, and by other non-coding elements of the genome. See also supplement Fig. S5.
