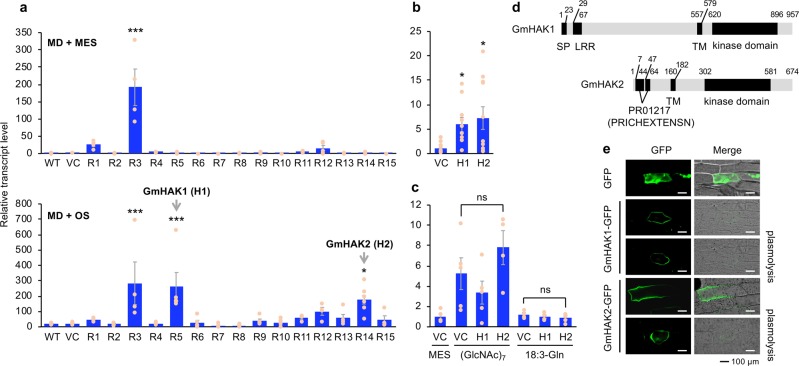
Fig. 1. Mining of GmHAKs and their structure and subcellular localization.
Transcript levels of PDF1.2 in leaves of wild-type (WT) plants, vector control (VC) plants, and 15 lines of GmHAK-expressing Arabidopsis plants (R1-R15) at 24 h after mechanical damage (MD) with application of MES buffer or OS from Spodoptera litura (a, n = 4–8). Moreover, VC, H1 (R5), and H2 (R14) lines were damaged with S. litura larvae (b, n = 10–11) or treated with MD + MES buffer, chitin oligosaccharides [(GlcNAc)7], or N-linolenoyl-l-Gln (18:3-Gln) (c, n = 4–5) for 24 h, and transcript levels of PDF1.2 in their leaves were measured. All the individual data points are shown with the means and standard errors. Data marked with an asterisk are significantly different from those of WT (a) or VC (b), based on an ANOVA with Holm’s sequential Bonferroni post hoc test (***P < 0.001; *0.01 ≤ P < 0.05). ns, not significant. d GmHAK1 and GmHAK2 proteins are schematically represented. LRR, leucine-rich repeat; SP, signal peptide; PR01217 (PRICHEXTENSN), proline-rich sequence; TM, transmembrane domain. e Subcellular localization of GmHAKs. The vector containing the CaMV 35S promoter (35SP)::GFP or 35SP:: GmHAK1 or GmHAK2 fused to GFP (GmHAK1-GFP or GmHAK2-GFP, respectively) was transformed into the epidermal cells of onions. Plasmolysis was induced by 0.6 M mannitol for 15 min. The photograph with GFP signal alone and the photograph with merged GFP signal and bright field images are shown.