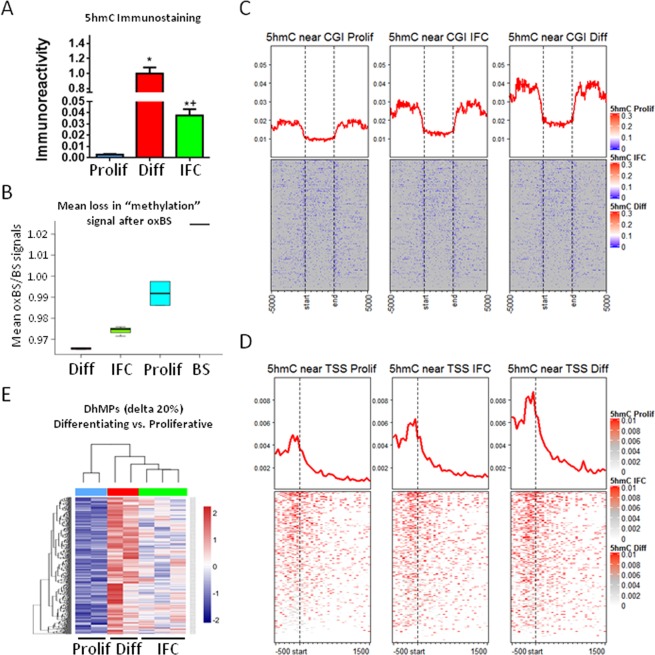
Figure 2.
5hmC genome-wide enrichment during hepatocyte differentiation. (A) Immunofluorescence of 5hmC in proliferative (Prolif), differentiating (Diff) and differentiating + IFC-305 (IFC) HepaRG cells. Quantification barplots of 5hmC signal are based on mean ± SEM from 3 fields per group. *Statistical difference (p < 0.05) compared with proliferative, or *+(p < 0.05) compared with proliferative and differentiating cells. (B) The same conditions were assessed for base-resolution 5mC/5hmC, using BS and OxBS followed by EPIC beadarray hybridization (see Methods). Proportion of signal loss after oxidation (mean oxBS signal / mean BS signal) was used as an estimation of global 5hmC in each condition. BS represents the variation between two BS technical replicates. Global distribution of 5hmC for one representative sample of each condition, according to CpG islands (CGI) (C) and transcription start sites (TSS) (D). In both cases, 5hmC levels are averaged across all hg19-annotated genomic regions. (E) Heatmap showing hydroxymethylome comparison between differentiating and proliferative cells. Differentially hydroxymethylated positions (DhMPs) were filtered by the magnitude of change in methylation (delta beta) of at least 20% and p-adjusted value <0.05. Two independent cultures were used for proliferative and differentiating cells, and three independent cultures for differentiating + 1 mM IFC-305.