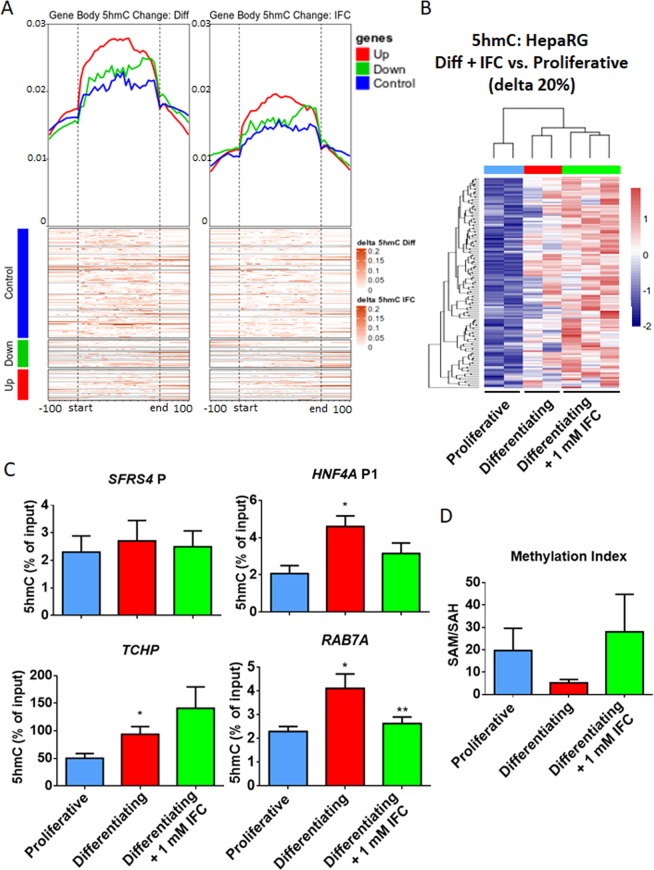
Figure 5.
Reduced 5hmC enrichment under methyl-donor perturbation. 5hmC in proliferative, differentiating and differentiating + IFC-305 HepaRG cells, as assessed using BS and OxBS followed by EPIC beadarray hybridization (see methods and Fig. 2). (A) Metagene heatmaps showing the shift of 5hmC content upon one week of HepaRG differentiation in control conditions (left panel) or after exposure to the adenosine derivative IFC-305 (right panel). The increase in 5hmC content is plotted separately for the bodies of genes known to be up- (red) or down- (green) regulated after one week of differentiation. Housekeeping genes that did not display any significant change are shown in blue. (B) Heatmap showing hydroxymethylome comparison between differentiating + IFC-305 and proliferative cells. Differentially hydroxymethylated positions (DhMPs) were filtered by the magnitude of change in methylation (delta beta) of at least 20% and p-adjusted value < 0.05. Two independent cultures were used for proliferative and differentiating cells, and three independent cultures for differentiating + 1 mM IFC-305. (C) Validation of 5hmC changes. 5hmC enrichment was assessed by hMeDIP and qPCR; SFRS4 gene promoter was used as non-5hmC enriched control region, HNF4A promoter P1 was used as one week differentiation 5hmC enrichment positive control, data represent mean ± SEM 3 independent cultures/condition; *Statistical difference (p < 0.05) compared with proliferative cells. **Statistical difference (p < 0.05) compared with differentiating cells. (D) Content of S-adenosylhomocysteine (SAH) and S-adenosylmethionine (SAM) was assessed using HPLC in different experimental conditions and samples added with standards. Barplots indicate the methylation index, calculated as the SAM/SAH ratio, mean ± SEM from 4 cultures/condition.