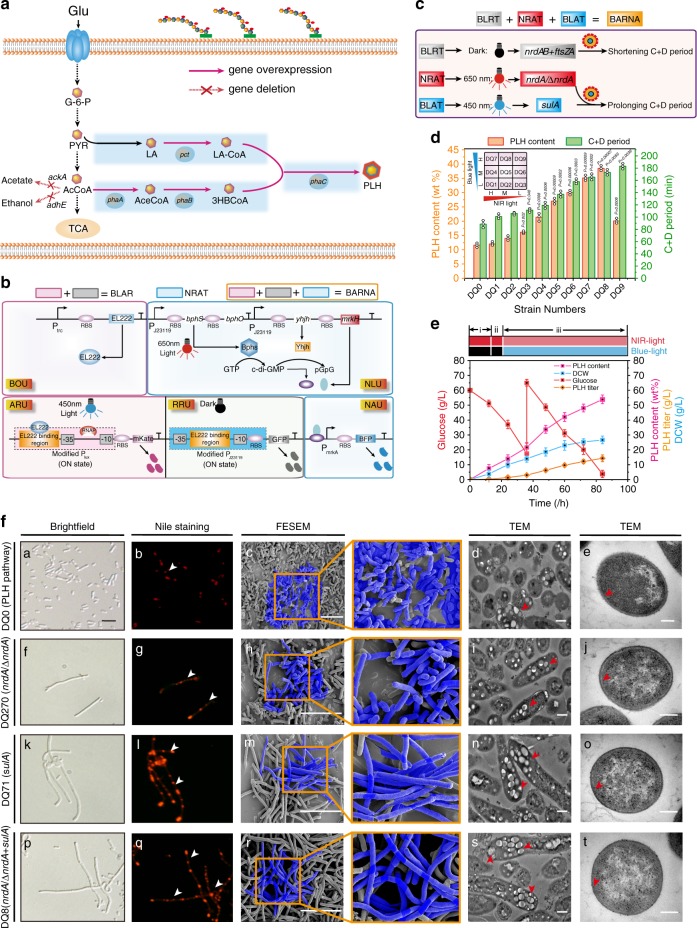
Fig. 6. Prolonging cell division for PLH production by the BARNA system.
a The schematic diagram of PLH biosynthesis pathway in E. coli DQ0. b The schematic diagram of BARNA containing BLRT and BANA. c Increasing PLH production by BARNA regulation. BLRT, BLAT, and NRAT were used to control nrdAB + ftsZA, sulA, and nrdA, respectively. d Effect of blue-light and NIR-light combination on PLH content and the C+D periods. H, M, and L were 0.8, 0.3, and 0.2 W/cm2, respectively (Supplementary Figs. 37A, B, 38A, B). The inserted figure was nine combination group (E. coli DQ1–DQ9) with orthogonally matrix by blue and NIR-light illumination intensity. e PLH production with the engineered E. coli DQ8 controlled by light stimulation during fed-batch fermentation. Phase I, II, and III were controlled by 0.8 W/cm2 NIR-light, 0.8 W/cm2 NIR-light, 0.8 W/cm2 blue-light and 0.3 W/cm2 NIR-light, respectively. f Effect of prolonging cell division on cell morphology during PLH production. The nrdA and sulA genes were constitutively expressed in E. coli DQ0, resulting in E. coli DQ270 and E. coli DQ271, respectively. The scale bar for c, h, m, r is 10 μm. The scale bar for d, i, n, and s is 0.5 μm. For e, j, o, and t is 0.2 μm. For a, f, k, and p is 5 μm. d, e values are shown as mean ± s.d. from three (n = 3) biological independent replicates. Glu glucose, G6P glucose 6-phosphate, PYR pyruvate, LA lactate, PEP phosphoenolpyruvate, AcCoA acetyl-CoA, aceCoA acetoacetyl-CoA, 3HBCoA 3-hydroxybutyryl-CoA, LAcoA lactyl-coA, ackA acetate kinase A, adhE alcohol dehydrogenase, phaA β-ketothiolase, phaB acetoacetyl-CoA reductase, phaC PHA synthase, pct propionyl-CoA transferase. Significance (p-value) was evaluated by two-sided t-test, compared to E. coli DQ0. Source data underlying Fig. 6d–f are provided as a Source data file.