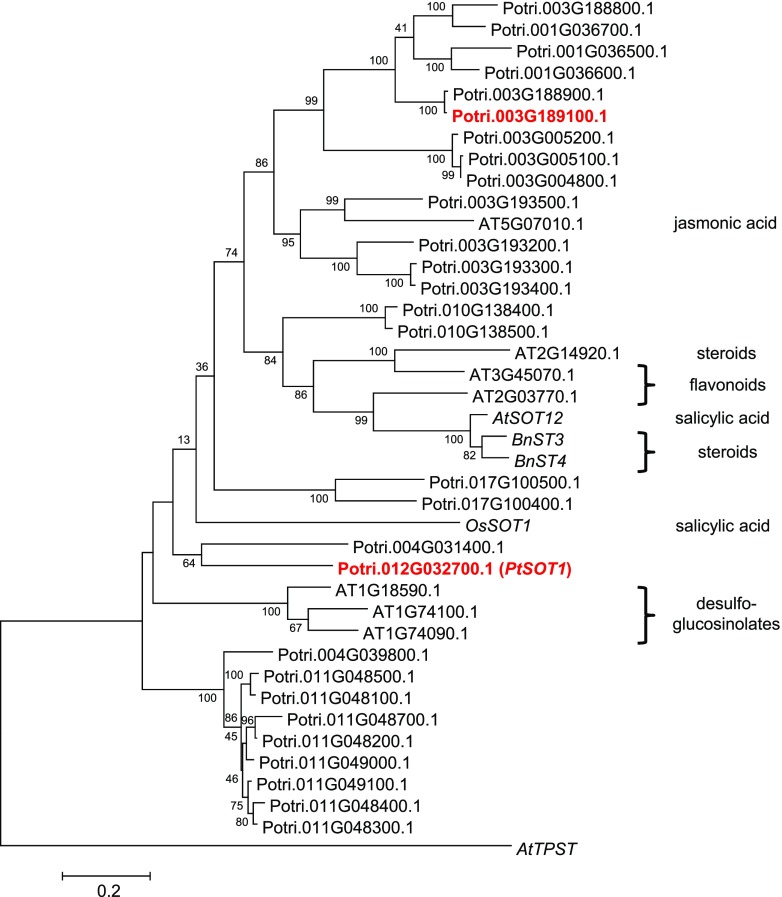
Figure 5.
Cladogram analysis of putative poplar SOT genes and characterized SOT genes from other plants. The tree was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model implemented in the software MEGA6 (Tamura et al., 2013). Bootstrap values (n = 1,000) are shown next to each node. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were first+second+third+Noncoding. All positions with <80% site coverage were eliminated. Arabidopsis AtTPST (NM_100677) was used as an outgroup. Poplar SOTs investigated in this study are highlighted in red. Substrates of characterized SOTs are shown next to the tree on the right side. AtSOT12, Arabidopsis NM_126423; BnST3, B. napus AF000307; BnST4, B. napus AY442306; OsSOT1, O. sativa LOC_Os11g30910.