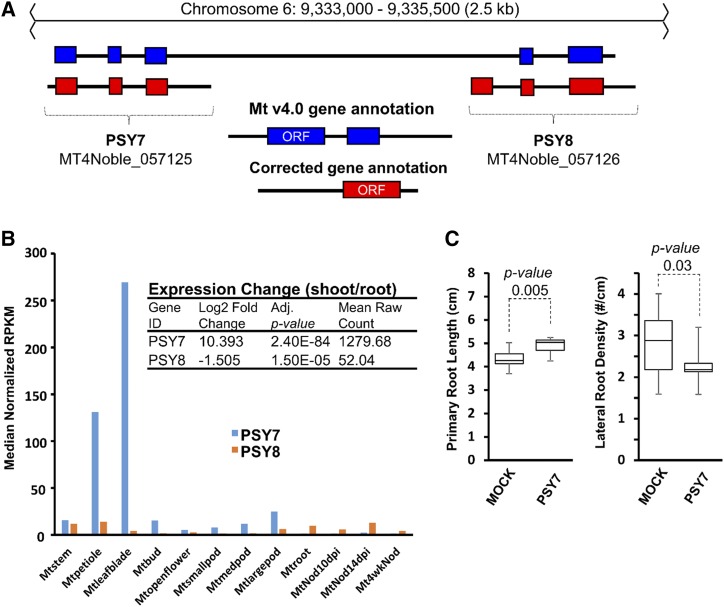
Figure 7.
Reannotation of a Plant Peptide Containing Sulfated Tyr (PSY) reveals a novel bioactive PSY SSP-coding gene. A, RNA-seq evidence indicates that Medtr6g027320.1 is annotated incorrectly in Mt v4.0. RNA-seq reads mapped to each locus are illustrated as a Sashimi plot representing the number of mapped reads at each nucleotide. Two distinct transcripts emerge when RNA-seq mapped reads are separated by expression in shoots or roots. Reads from one representative biological replicate are presented above. The new transcripts uncover an additional upstream PSY SSP-coding gene and have been renamed PSY7 and PSY8 accordingly. B, A plot of tissue expression levels of the newly uncovered PSY7 and PSY8 genes. PSY7 is the dominantly expressed transcript and is mostly expressed in leaves and shoots. In contrast, PSY8 demonstrates low-level expression throughout roots, nodules, and stems/petioles. The inset shows how expression changes from shoot versus root highlight the DE of the two PSY transcripts. RPKM, reads per kilobase of transcript per million mapped reads. C, Synthetic PSY7 peptide shows bioactivity when tested in the root screening platform, leading to enhanced primary root length (consistent with its characterized role in cell division/expansion), while also suppressing lateral root density. Plots show the median value (center lines), box limits indicate the 25th and 75th percentiles, and the bars indicate the maximum and minimum values among the data points collected (Students t test). n = 10 for peptide-treated plants and 20 for mock-treated plants.