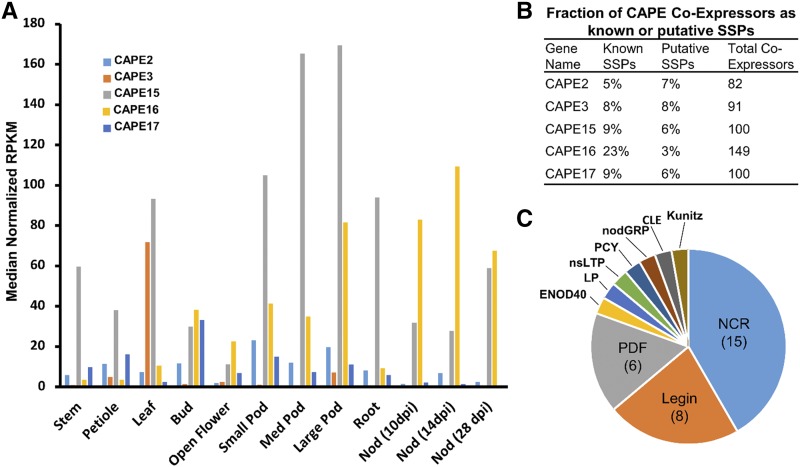
Figure 9.
CAPE16 expression predominates in symbiotic nodules and coexpresses with many SSPs. A, RNA-seq data from the MtSSPdb. Five CAPE peptides, which have been shown previously to be induced by macronutrient deficiencies, were interrogated for expression patterns across 12 different organs. Nod, Nodule; RPKM, reads per kilobase of transcript per million mapped reads. B, CAPE16 shows a strong enrichment in coexpressing SSP-coding genes. Coexpressing SSPs were defined as those with a Pearson correlation coefficient >0.8 across 122 different experimental conditions. Thirty-five of a total 149 coexpressors of CAPE16 (23%) were SSP genes, versus 5% to 9% of the other four CAPE genes. C, Pie chart showing the distribution of the CAPE16 coexpressing SSP-coding genes, most of which are NCR peptides, leginsulins (Legin), or plant defensins (PDF). ENOD40, Early Nodulin 40; LP, LEED.PEED; nsLTP, nonspecific lipid transfer protein; nodGRP, nodule-specific Gly-rich protein.