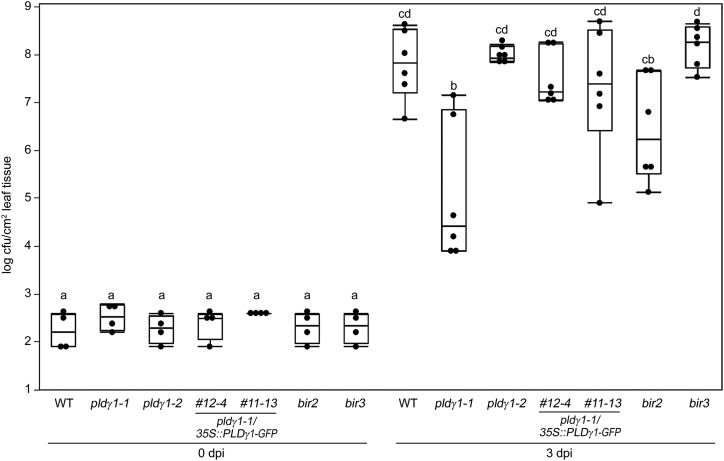
Figure 1.
Genetic inactivation of PLDγ1 results in increased resistance to bacterial infection. Wild-type (WT) plants or the indicated mutant lines for pldγ1-1, pldγ1-2, complementation lines 12-4 and 11-13 (pldγ1-1/35S:: PLDγ1-GFP, T4 generation), or bir2 and bir3 were infiltrated with 104 cfu/mL of virulent Pseudomonas syringae pv. tomato DC3000. At 0 and 3 d postinoculation (dpi), bacterial growth was quantified by counting colony-forming units. Box plots show the minimum, first quartile, median, third quartile, and a maximum of log cfu/cm2 leaf tissue (n = 4 for 0 dpi; n = 6 for 3 dpi). Lowercase letters indicate homogenous groups according to post hoc comparisons following one-way ANOVA (Tukey-Kramer multiple comparison analysis at a probability level of P < 0.05). The entire experiment was repeated three times with similar results.