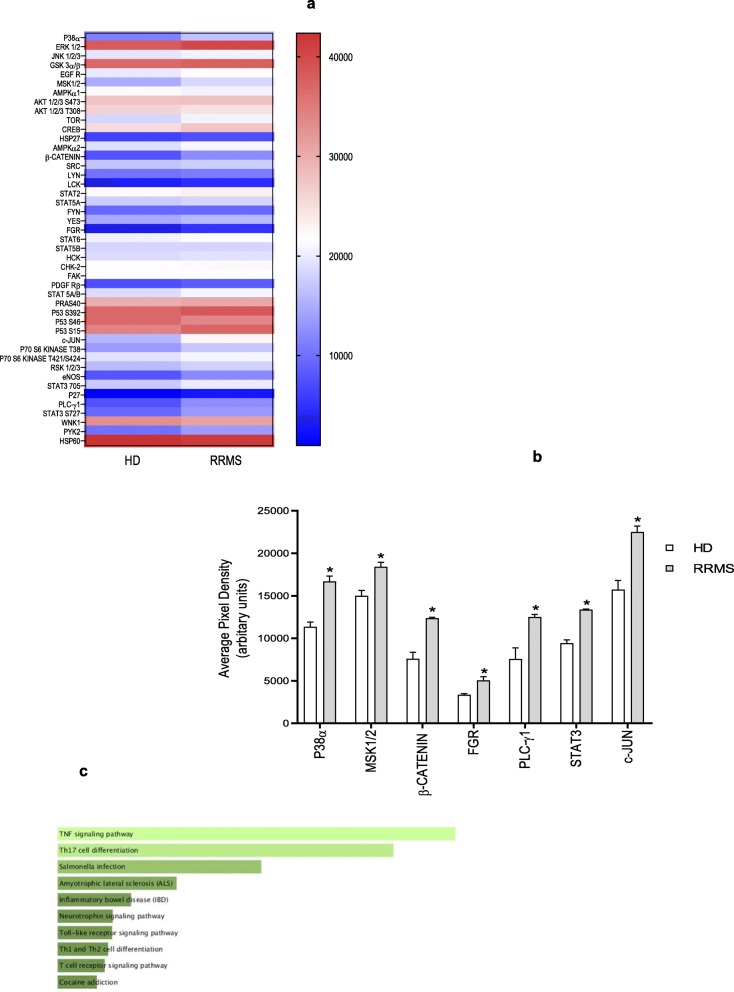
Fig. 9.
Phospho-protein activation. a The map display the expression of differentially expressed proteins identified from the proteomics analysis in hCMEC/D3 cells treated overnight with 20% RRMS naïve-to-treatment or HD sera performed on 10 donors pooled per group pooled (in sex, age and EDSS score for RRMS patients), in duplicate. The colour scale illustrates the expression level of each protein across the 2 samples; the colour intensity indicates the degree of protein up- or downregulation. b Average pixel density of phosphorylated proteins known to be involved in pro-inflammatory processes/pathways significantly different. c Association of all significantly phosphorylated enzymes with KEGG pathways, ranked in order of p value, Enrichr. Data are shown as mean ± s.e.m. (statistical analysis by Mann-Whitney U test (two tails) *P ≤ 0.05, **P ≤ 0.01