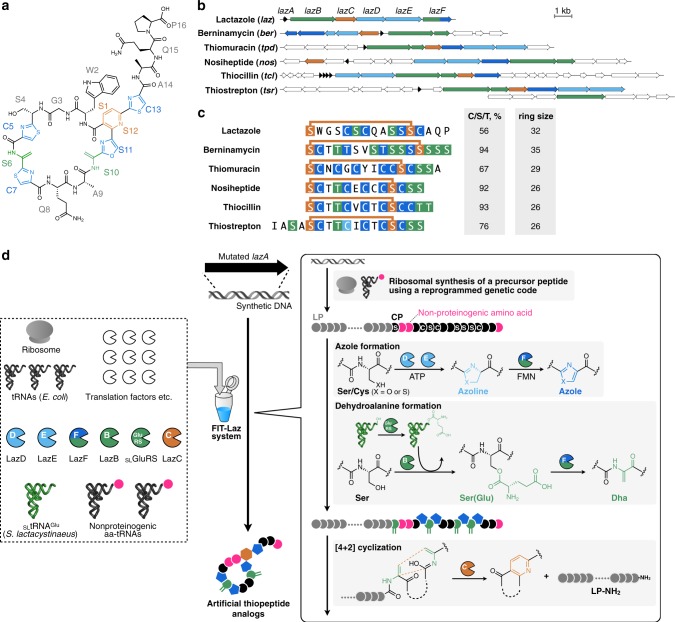
Fig. 1. Lactazole A and its biosynthesis with the FIT-Laz system.
a Chemical structure of lactazole A. b Comparison of laz BGC with other prototypical thiopeptide BGCs. Homologs of laz genes are color-coded. Genes encoding enzymes responsible for the installation of azolines, azoles, dehydroalanine, and pyridine are shown in light blue, blue, green, and orange, respectively. Precursor peptide structural genes are shown in black and ancillary genes absent from laz BGC are in white. c Comparison of primary sequences for thiopeptides from b, with the same PTM color coding. The comparison reveals an unusual macrocycle size, low C/S/T content, and the absence of azole modification in position 2 as unique features of lactazole. d Summary of the FIT-Laz system and the roles of individual enzymes during lactazole biosynthesis. In FIT-Laz, synthetic DNA templates encoding LazA or its mutants are in-vitro transcribed and translated to generate precursor peptides, which undergo a cascade of PTMs introduced by lactazole biosynthetic enzymes to yield lactazole A or its artificial analogs.