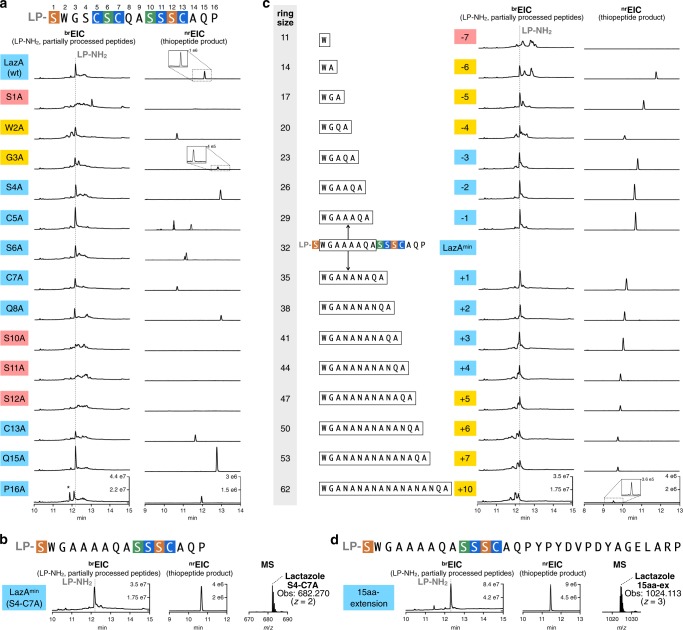
Fig. 3. Substrate scope of the FIT-Laz system.
a Ala scanning of the LazA CP. Single-point Ala mutants of LazA were treated with the full enzyme set and the outcomes were analyzed by LC-MS. Displayed are LC-MS chromatograms (brEIC chromatograms on the left showing partially processed linear peptides and LP-NH2 after enzymatic treatment, and nrEIC chromatograms on the right for expected thiopeptides generated at m/z 0.10 tolerance window). For mutants highlighted in light blue biosynthesis proceeded efficiently; yellow highlighting indicates inefficient thiopeptide formation accompanied by the accumulation of linear intermediates and side products; red indicates mutants that failed to yield a detectable thiopeptide. Peaks denoted with an asterisk (*) indicate translation side products. Mutants C5A and S6A gave 4 and 2 thiopeptides, respectively, annotations of which can be found in Supplementary Figs. 16 and 17. Y-axes are scaled between samples for each chromatogram type. b LC-MS chromatograms as in a for the enzymatic processing of LazAmin on the left with a zoomed-in mass spectrum of the produced thiopeptide on the right. c LC-MS chromatograms as in a for ring expansion and contraction study of LazAmin. d LC-MS chromatograms and mass spectrum as in b for a LazAmin variant containing a 15-amino acid extension in the tail region. Collectively, these data point to remarkable substrate tolerance of Laz enzymes.