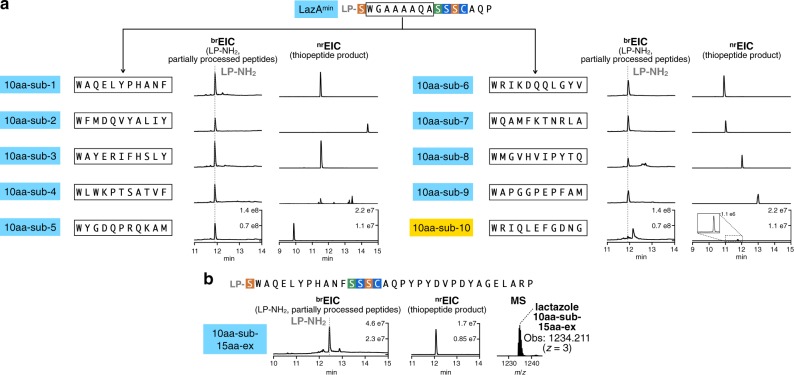
Fig. 4. Synthesis of lactazole-like thiopeptides with randomized sequences.
a LazAmin variants containing ten consecutively randomized amino acids were first treated with LazDEF and then with LazBC, tRNAGlu, and GluRS (see Methods for details), and the outcomes were analyzed by LC-MS. Displayed are LC-MS chromatograms (brEIC on the left showing partially processed linear peptides and LP-NH2 after enzymatic treatment, and nrEIC chromatograms on the right for expected thiopeptides generated at m/z 0.10 tolerance window). Mutants highlighted in light blue indicate efficient thiopeptide assembly; in yellow, inefficient thiopeptide formation accompanied by the accumulation of linear intermediates and side products. One construct, 10aa-sub4, resulted in eight different thiopeptides, partial annotation of which can be found in Supplementary Fig. 20. Efficient in-vitro biosynthesis observed in nine out of ten cases underscores the substrate plasticity of FIT-Laz. b LC-MS chromatograms as in a for the enzymatic processing of a 34 amino acid-long LazAmin variant on the left with a zoomed-in mass spectrum of the produced thiopeptide on the right.