Fig. 2. Multiple Tup1–Cyc8 interacting TFs associate with the IME1 promoter.
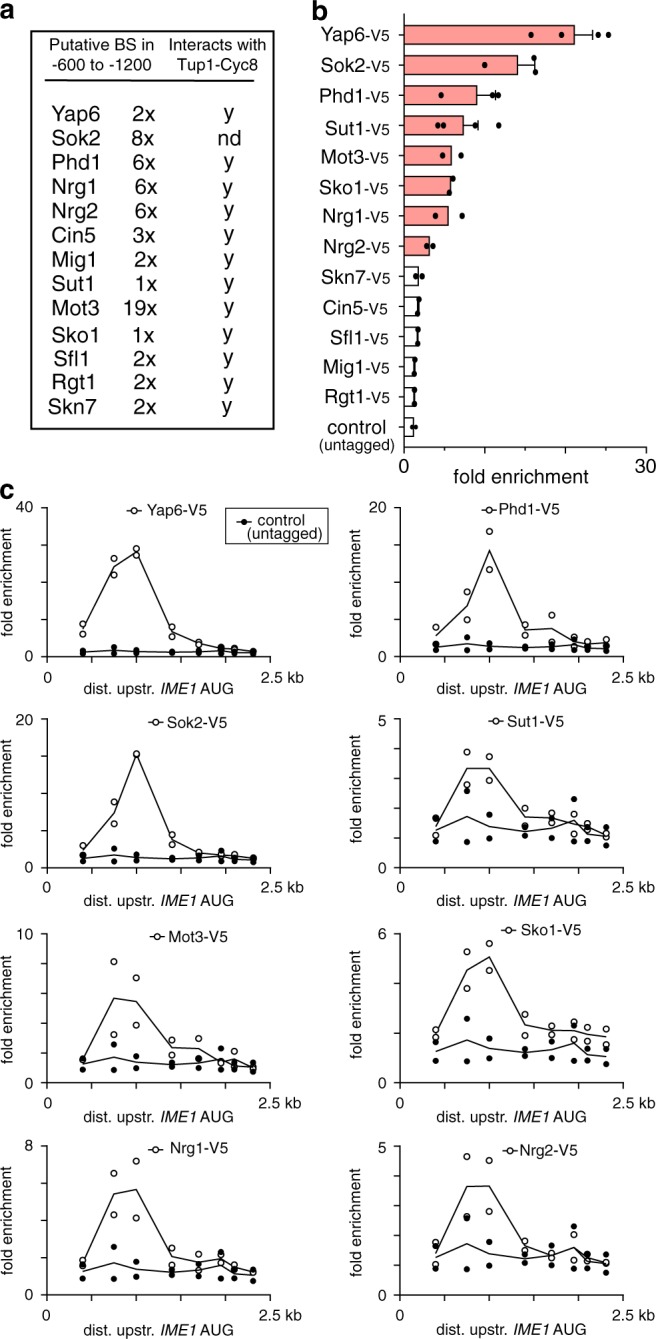
a Table of TFs that have been implicated to interact with Tup1–Cyc8 and have motif sequences within the IME1 promoter. BS = binding site. b Multiple TFs associate with the IME1 promoter in the same region where Tup1–Cyc8 binds. TFs listed in a were tagged with V5 epitope tag. Diploid cells harbouring V5-tagged transcription factor (YAP6-V5, FW3833; SOK2-V5, FW5638; PHD1-V5, FW4466; SUT1-V5, FW6974; MOT3-V5, FW4383; SKO1-V5, FW4389; NRG1-V5, FW4393; NRG2-V5, FW4396; SKN7-V5, FW4399; CIN5-V5, FW7072; SFL1-V5, FW7070; MIG1-V5, FW4665; RGT1-V5, FW4386) and control cells (untagged, FW1511) were grown till exponential growth. The region around 1000 bp upstream of IME1 AUG was analysed for binding by ChIP. The signals were normalised over HMR. Red bars represent a signal of three-fold over HMR or more. Mean and SEM of n = 4 (Yap6-V5 and Sut1-V5), n = 3 (Sok2-V5 and Phd1-V5) and n = 2 for other TFs plus untagged control are shown. c Similar as b, except that TFs that showed at least three-fold enrichment in b were analysed for binding throughout the whole IME1 promoter. Eight primer pairs were used for the analyses. The mean of n = 2 is shown.
