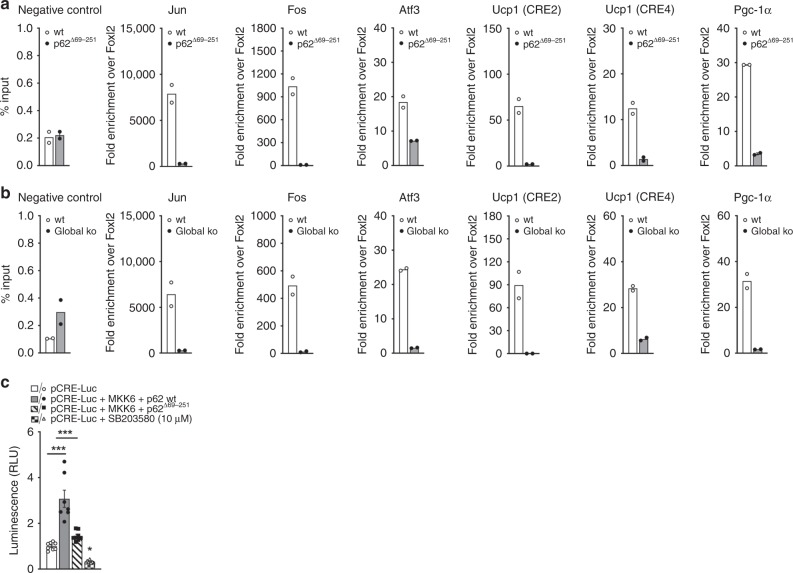
Fig. 7. Impaired genomic binding of ATF2 in p62Δ69-251 and global p62−/− mice.
Chromatin immunoprecipitation (ChIP) in BAT of male wt and p62Δ69-251 mice (a) and p62−/− mice (b) using antibodies specific for ATF2. ChIP-qPCR analysis of Foxl2 (negative control), Jun, Fos, Atf3, Ucp1 (CRE2), Ucp1 (CRE4), and Pgc-1α (n = 5 mice pooled each genotype, n = 2 technical replicates) (a, b). Luciferase-based CRE reporter assay of HEK293T cells transfected with the pCRE-Luc reporter, a dominant positive MKK6 (K82A) and p62 wt or p62Δ69-251 (n = 7/8 technical replicates each group) (c). Panel (c) is a representative example of three independently performed studies, each yielding similar results. Data in (a) and (b) have been analyzed using a two-sided two-tailed t-test, data in (c) have been analyzed using one-way ANOVA was treatment as co-variant and using Bonferroni multiple comparison between the treatment groups. Data represent mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001. Exact p-values are (c) pCRE-Luc vs. pCRE-Luc + MKK6 + p62 wt p < 0.0001, pCRE-Luc vs. pCRE-Luc + MKK6 + p62Δ69-251 p > 0.05, pCRE-Luc vs. pCRE-Luc + MKK6 + SB203580 p > 0.05, pCRE-Luc + MKK6 + p62 wt vs. pCRE-Luc + MKK6 + p62Δ69-251 p < 0.0001, pCRE-Luc + MKK6 + p62 wt vs. pCRE-Luc + MKK6 + SB203580 p > 0.0001, pCRE-Luc + MKK6 + p62Δ69-251 vs. pCRE-Luc + MKK6 + SB203580 p = 0.0005. Foxl2 forkhead box L2, Fos FBJ osteosarcoma oncogene, Atf3 activating transcription factor 3.