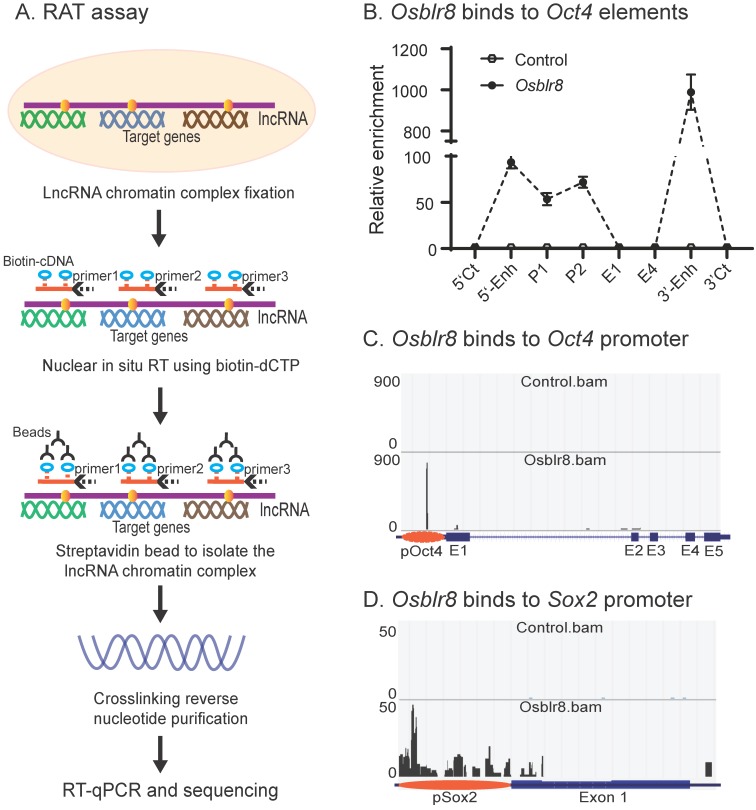
Figure 6.
Mapping the Osblr8 interaction network by RAT-seq. A) RNA reverse transcription-associated trap (RAT) assay. After fixation, Osblr8 lncRNA was reverse transcribed into cDNAs in the isolated nuclei with biotin-dCTP using three gene specific reverse primers. The DNA-protein-lncRNA-biotin-cDNA complex was further purified from genomic DNAs by biotin-streptavidin beads. After the crosslinking reversal, the captured DNAs were used for Illumina library sequencing or analyzed by Q-PCR to identify the Osblr8 interaction network. B) Osblr8 lncRNA-Oct4 DNA interactions by Q-PCR. RAT library samples were used to perform Q-PCR to quantitate binding intensity. The results were normalized to the value of the streptavidin bead pulldown control. Control: the RAT-seq library was constructed using random oligo primers; Obelr20: the RAT-seq library was constructed using Osblr8-speccific primers; 5'-CT, 3'-CT: the RAT 5'- and 3'- control sites; P: promoter; E1 and E4: Oc4 exons 1 and 5. C) Interaction of Osblr8 at the Oct4 locus. Control: the RAT library was constructed with random oligo primers. Osblr8: the RAT library was constructed using Osblr8 complementary primers; 5'-Enh: 5'-enhancer; E1-E5: Oct4 exons 1-5; 3'-Enh: 3'-enhancer. Note the enriched binding of Osblr8 lncRNA at the Oct4 promoter. D) Interaction of Osblr8 at the Sox2 locus. Osblr8 binds to the Sox2 promoter area.