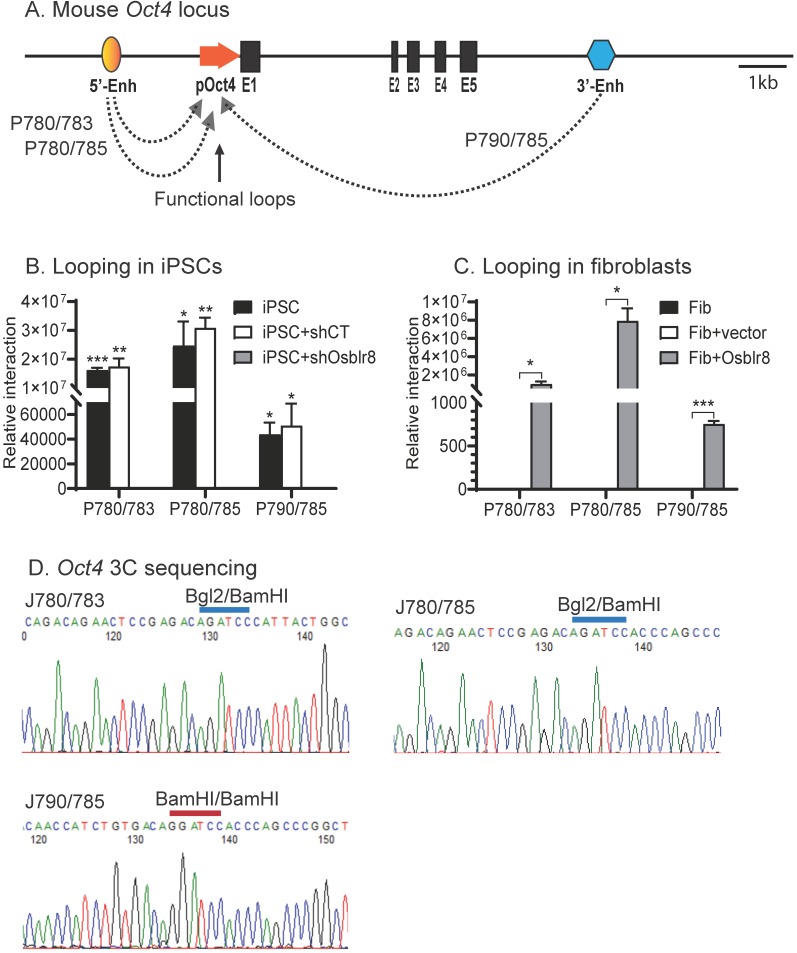
Figure 7.
Osblr8 orchestrates pluripotency-specific intrachromosomal looping. A) Location of 3C primers used to detect the interaction between the Oct4 promoter and enhancer. Enh: enhancers; pOct4: Oct4 promoter; E1-E5: exons; Arrows: intrachromosomal interactions. B) Knockdown of Osblr8 abolishes the intrachromosomal interaction loop. shCT: negative control shRNA, shOsblr8: shRNA that targets Osblr8 lncRNA. iPSC: induced pluripotent stem cells. Primer sets that detect the presence of looping are marked in Fig.7A. The 3C interaction was quantitated by qPCR and was standardized over the 3C control Ercc3 gene. For comparison, the relative 3C interaction was calculated by setting the 5' or 3' control as 1. *P<0.05, ** P< 0.01 as compared with the shOsblr8 treatment and shRNA control. C) Overexpression of Osblr8 induces de novo formation of intrachromosomal looping. FIB: fibroblasts. Fib+Vector: fibroblasts transfected with vector control; Fib+Osblr8: fibroblasts overexpressed Osblr8. D) Sequencing of the Oct4 intrachromosomal loop products. Blue line on the top of the sequence: the 3C ligation product between the BamH1 and Bgl2 sites. Red line on the top of the sequence: the 3C ligation product between the BamH1 and BamH1 sites.