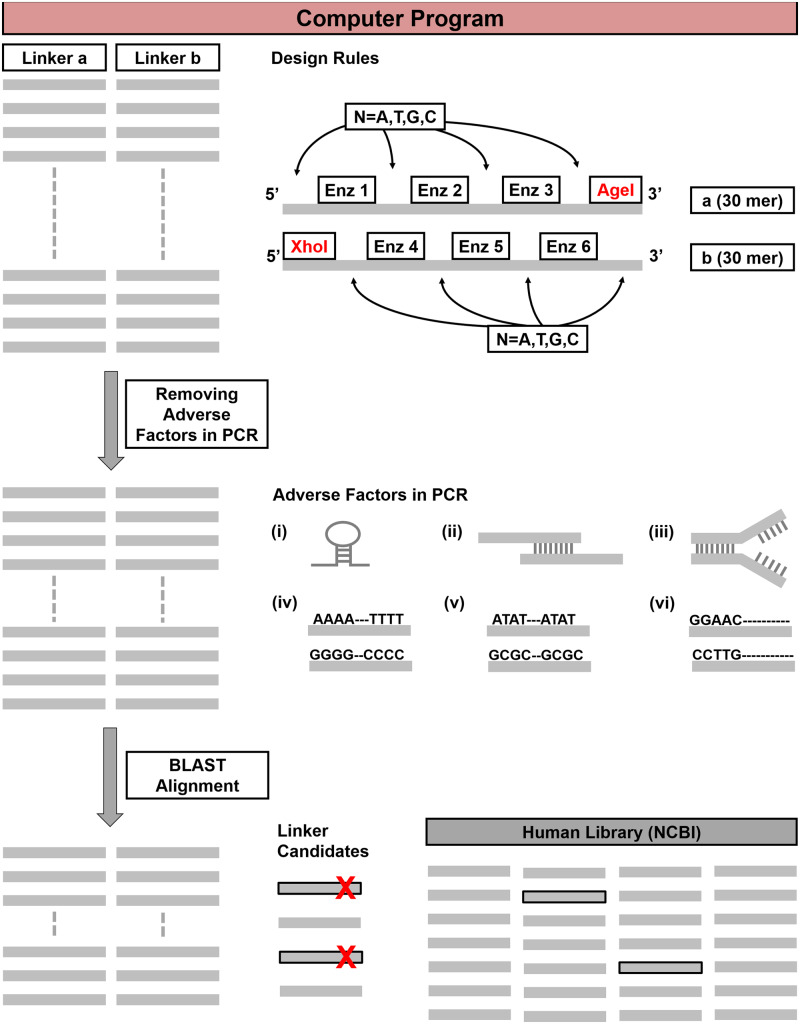
Figure 1:
In silico screening of the linkers. We developed a computational pipeline to design the linker pairs with the length of 30 bps, containing specific RESs. For a linker pair (linkers a and b), there were total eight RESs assigned. AgeI site was at the 5’ end of linker a, XhoI was at the 3’ end of linker b, and six other RESs (Enzs 1–6) in the middle. “N” represents any nucleotides localized between the RESs for filling up the spaces and creating the probability of the sequence combinations for the linkers. According to our concept, as the first step, the computer program generated a list of all the possible linker pair sequences. Next, the sequences that harbor adverse factors for PCR reaction were removed. Examples for adverse factors are (i) single-stranded DNA secondary structures, such as hairpin, (ii) potential of dimerization, (iii) high salt-adjusted melting temperature, (iv) unbalanced GC content, (v) nucleotide repeats, and (vi) GC clamp. Last, the BLAST alignment step was applied to further filter the candidates and select the linker pairs with low identity to the human genome.