Figure 3:
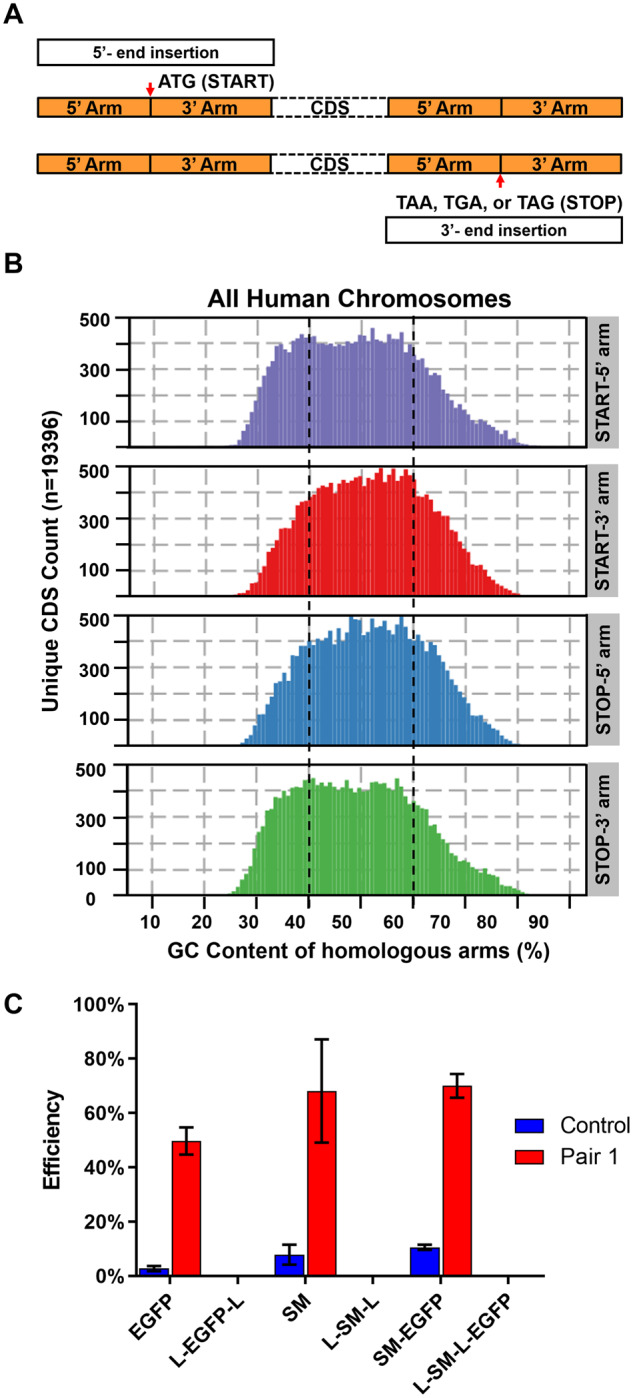
Unbalanced GC content and removable SM were the unfavorable factors affecting Gibson assembly. (A) The outline of the insertion sites for a target gene. The red arrows show the insertion positions of a gene. (B) The genome-wide distribution of the GC content for the 750 bps sequences at both sides of the start/stop codon. The unbalanced GC content is defined as higher than 60% or lower than 40% (black dashed lines). (C) The efficiency of Gibson assembly reaction for the IF with/without LoxP sites. Three different IFs, EGFP, SM and SM-EGFP were assembled with the homologous arms of CDH1 and the IF vector. For the IF vector, the control vector and the one containing linker pair 1 were applied. The efficiency was calculated as the number of the colonies with the desirable constructs divided by the number of tested colonies. The experiments were conducted in triplicate. The error bars indicate mean ± SD. L, LoxP.
