Figure 4. Comparison between Rap1 and Ras.
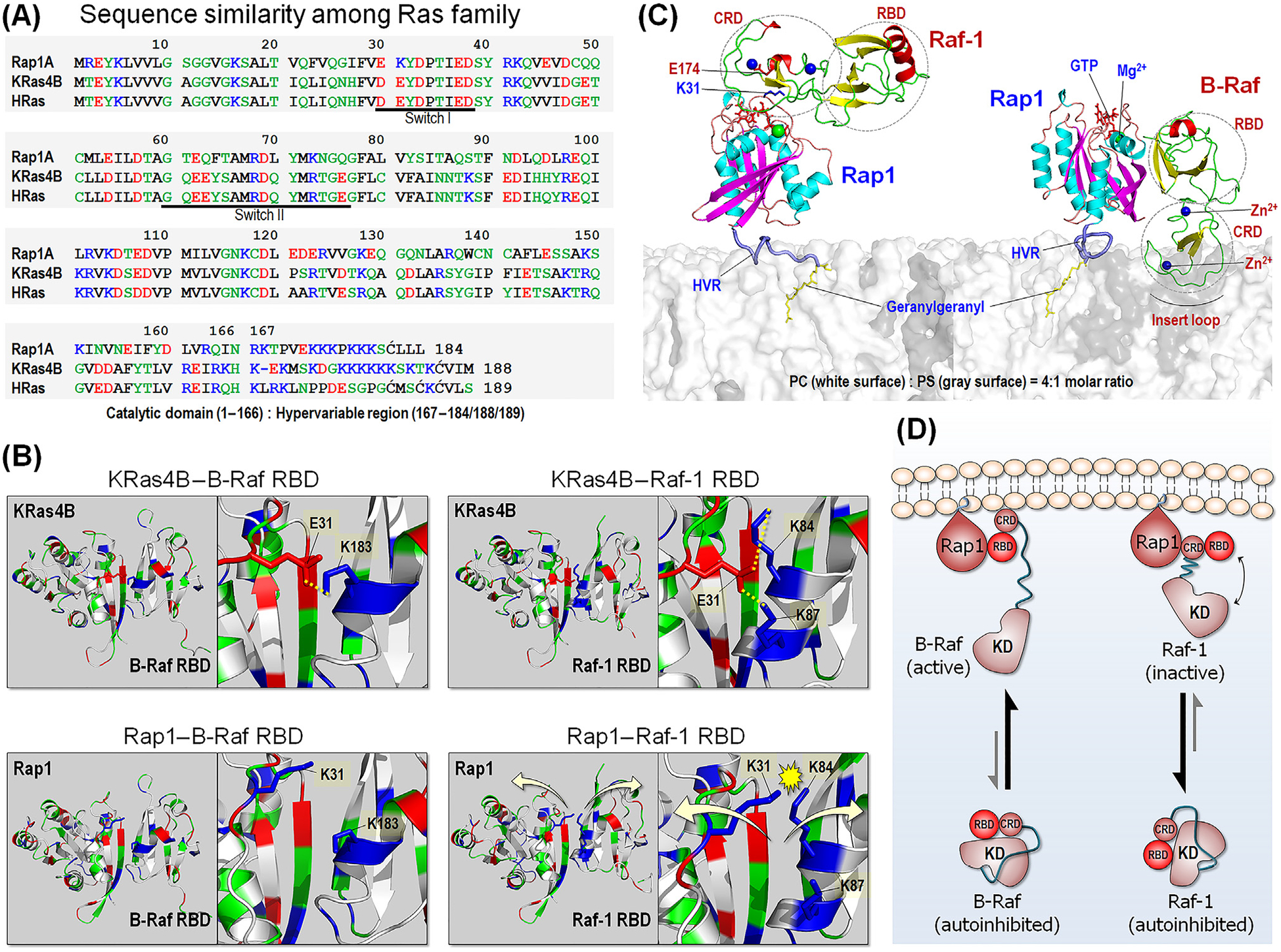
(A) Sequence similarity among Ras family, including Rap1, KRas4B, and HRas. In the sequence, hydrophobic, polar/glycine, positively charged, and negatively charged residues are colored black, green, blue, and red, respectively. (B) Model structures of KRas4B catalytic domain interacting with BRAF Ras-binding domain (RBD) (upper left) and Raf-1 RBD (upper right), based on our previous studies [87]. In addition to the strong β-sheet interaction, Ras adds stability to the interaction with the RBDs through the salt bridges between E31 and K183 for BRAF, and between E31 and K84/K87 for Raf-1. Rap1 can interact with BRAF RBD due to distant repulsive force between K31 and K183 (lower left). However, in the interaction with Raf-1 RBD, Rap1 K31 electrostatically crashes with K84/K87, leading to dissociation. (C) Model structures of Rap1 interacting with Raf-1 conserved region (CR)1 (left) and BRAF CR1 at the anionic membrane. Rap1 prefers to bind Raf-1 cysteine-rich domain (CRD), while it binds to BRAF RBD. (D) Schematic diagram of Raf activation by Rap1. Abbreviations: KD, kinase domain; HVR, hypervariable region.
