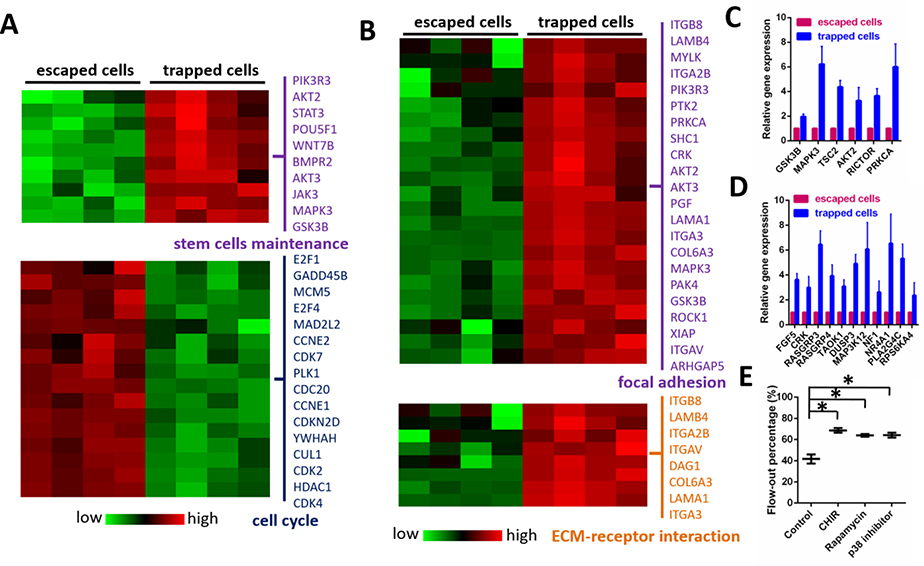
Figure 4. Sorted HSPCs exhibit differences in gene expression associated with the regulation of HSPC mobilization and maintenance.
(A) RNA-seq analysis was used to generate a heat map demonstrating expression levels of genes associated with the regulation of stem-cell maintenance and cell-cycle progression in trapped and escaped HSPCs. (B) RNA-seq analysis was used to generate a heat map showing the expression levels of genes mediating focal adhesion and ECM-receptor interactions in trapped and escaped HSPCs. (C) Expression of genes associated with GSK3 and mTOR pathways was analyzed in trapped and escaped HSPCs. Data are presented as mean ± s.d. from three different experiments (n=3, measured three times, for each group). (D) Expression of genes associated with MAPK signaling was analyzed in trapped and escaped HSPCs. Data are presented as mean ± s.d. from three different experiments. (n=3, measured three times, for each group) (E) Percentage of HSPCs escaping the traps and flowing out of the chip in the presence or absence of indicated inhibitors. CHIR (1 μM), Rapamycin (1 μM) and p38 inhibitor (1 μM) were used to inhibit GSK3, mTOR and MAPK signaling pathways respectively. For each experiment, a total of ~150 cells in each device were analyzed to calculate the flow-out percentage. Data are presented as mean ± s.d. from three different experiments (n=3, measured three times, for each group). *P < 0.01 determined by a two-tailed Student’s t test.