Summary
The unprecedented pandemic of pneumonia caused by a novel coronavirus, SARS-CoV-2, in China and beyond has had major public health impacts on a global scale [1, 2]. Although bats are regarded as the most likely natural hosts for SARS-CoV-2 [3], the origins of the virus remain unclear. Here, we report a novel bat-derived coronavirus, denoted RmYN02, identified from a metagenomic analysis of samples from 227 bats collected from Yunnan Province in China between May and October 2019. Notably, RmYN02 shares 93.3% nucleotide identity with SARS-CoV-2 at the scale of the complete virus genome and 97.2% identity in the 1ab gene, in which it is the closest relative of SARS-CoV-2 reported to date. In contrast, RmYN02 showed low sequence identity (61.3%) to SARS-CoV-2 in the receptor-binding domain (RBD) and might not bind to angiotensin-converting enzyme 2 (ACE2). Critically, and in a similar manner to SARS-CoV-2, RmYN02 was characterized by the insertion of multiple amino acids at the junction site of the S1 and S2 subunits of the spike (S) protein. This provides strong evidence that such insertion events can occur naturally in animal betacoronaviruses.
Keywords: SARS-CoV-2, bat coronavirus, COVID-19, spike protein, S1/S2 cleavage site
Highlights
-
•
Metagenomic analysis identified a novel coronavirus, RmYN02, from R. malayanus
-
•
RmYN02 was the closest relative of SARS-CoV-2 in most of the virus genome
-
•
Two loop deletions in RBD may reduce the binding of RmYN02 with ACE2
-
•
RmYN02 contains an insertion at the S1/S2 cleavage site in the spike protein
Zhou et al. report a bat-derived coronavirus, RmYN02, which is the closest relative of SARS-CoV-2 in most of the virus genome reported to date. RmYN02 contains an insertion at the S1/S2 cleavage site in the spike protein in a similar manner to SARS-CoV-2. This suggests that such insertion events can occur naturally in animal betacoronaviruses.
Results and Discussion
Coronaviruses (CoVs) are common viral respiratory pathogens that primarily cause symptoms in the upper respiratory and gastrointestinal tracts. In 1960s, two CoVs, 229E and OC43, were identified in clinical samples from patients experiencing the common cold [4]. More recently, four additional human CoVs have been successively identified: severe acute respiratory syndrome coronavirus (SARS-CoV) in 2002, NL63 in late 2004, HKU1 in January 2005, and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012. However, two betacoronaviruses (beta-CoVs), SARS-CoV and MERS-CoV, are notable as they have caused severe and fatal infections, leading to 774 and 858 deaths to date, respectively, suggesting that beta-CoVs may be of particular concern to human health. In December 2019, viral pneumonia caused by an unidentified microbial agent was reported, which was soon identified to be a novel coronavirus [1, 2, 3], now termed SARS-CoV-2 [5]. The number of patients infected with SARS-CoV-2 has increased sharply since January 21, 2020, and confirmed SARS-CoV-2 cases were present in all the Chinese provinces and municipalities by the end of January. The virus also spread rapidly outside of China in March and the World Health Organization had to declare a coronavirus disease 2019 (COVID-19) pandemic on March 11, 2020. As of April 15, 2020, more than two million confirmed SARS-CoV-2 cases have been reported, with >130,000 deaths worldwide.
An epidemiological survey of several SARS-CoV-2 cases at an early stage of the outbreak revealed that most had visited the Huanan seafood market in Wuhan city prior to illness, where various wild animals were on sale before it was closed on January 1, 2020, due to the outbreak. Phylogenetic analysis has revealed that SARS-CoV-2 is a novel beta-CoV distinct from SARS-CoV and MERS-CoV [1, 2, 3]. To date, the most closely related virus to SARS-CoV-2 is RaTG13, identified from a Rhinolophus affinis bat sampled in Yunnan province in 2013 [3]. This virus shared 96.1% nucleotide identity and 92.9% identity in the S gene, again suggesting that bats play a key role as coronavirus reservoirs [3]. Notably, however, several novel beta-CoVs related to SARS-CoV-2 have also been identified in Malayan pangolins (Manis javanica) that were illegally imported into Guangxi (GX) and Guangdong (GD) provinces, southern China [6, 7, 8]. Although these pangolin CoVs are more distant to SARS-CoV-2 than RaTG13 across the virus genome as a whole, they are very similar to SARS-CoV-2 in the receptor-binding domain (RBD) of the S protein, including at the amino acid residues thought to mediate binding to ACE2 [7]. It is therefore possible that pangolins play an important role in the ecology and evolution of CoVs. Indeed, the discovery of viruses in pangolins suggests that there is a wide diversity of CoVs still to be sampled in wildlife, some of which may be directly involved in the emergence of SARS-CoV-2.
Between May and October 2019, we collected a total of 302 samples from 227 bats from Mengla County, Yunnan Province in southern China (Table S1). These bats belonged to 20 different species, with the majority of samples from Rhinolophus malayanus (n = 48, 21.1%), Hipposideros larvatus (n = 41, 18.1%), and Rhinolophus stheno (n = 39, 17.2%). The samples comprised multiple tissues, including patagium (n = 219), lung (n = 2) and liver (n = 3), and feces (n = 78). All but three bats were sampled alive and subsequently released. Based on the bat species primarily identified according to morphological criteria and confirmed through DNA barcoding, the 224 tissues and 78 feces were merged into 38 and 18 pools, respectively, with each pool including 1 to 11 samples of the same type (Table S1). These pooled samples were then used for next-generation sequencing (NGS).
Using next-generation metagenomic sequencing, we successfully obtained 11,954 and 64,224 reads from pool 39 (from a total of 78,477,464 clean reads) that mapped to a previously identified SARS-like bat coronavirus, Cp/Yunnan2011 [9] (JX993988), and to SARS-CoV-2. From this, we generated two preliminary consensus sequences. After a series of verification steps, including re-mapping, one partial (23,395 bp) and one complete (29,671 bp) beta-CoV genome sequences were obtained and named BetaCoV/Rm/Yunnan/YN01/2019 (RmYN01) and BetaCoV/Rm/Yunnan/YN02/2019 (RmYN02), respectively. Pool 39 comprised 11 fecal samples from Rhinolophus malayanus collected between May 6 and July 30, 2019. RNAs from eight samples remained for further analysis. Based on the assembled sequence of RmYN02, TaqMan-based qPCR was performed to detect the existence of RmYN02 in the eight additional samples (see primers and probe in Table S2): the virus was only detected in sample no. 123 collected on June 25, 2019, from Rhinolophus malayanus with a cycle threshold (Ct) value of 34.76 (Figure S1). RmYN01 and RmYN02 were also identified in sample 123 using Tip green supermix (TransGen), with the Ct values 32.21 for RmYN02 and 35.54 for RmYN01. Sample 123 was therefore used to verify the RmYN02 genome sequence using the RmYN02-specific primers (Table S2). The partially amplified gene fragments were ∼10,000 bp in length covering the spike (Figure S2A) and 1b genes of RmYN02, and results from Sanger sequencing were consistent with those from metagenomic sequencing. Notably, 20 positions in the RmYN02 genome displayed nucleotide polymorphisms in the NGS data, although these did not include the S1/S2 cleavage site (Figure S2B). Only a few reads in the remaining 55 pools could be mapped to the reference CoV genomes. The sequence identity between RmYN01 and Cp/Yunnan2011 across the aligned regions was 96.9%, whereas that between RmYN01 and SARS-CoV-2 was only 79.7% across the aligned regions and 70.4% in the spike gene.
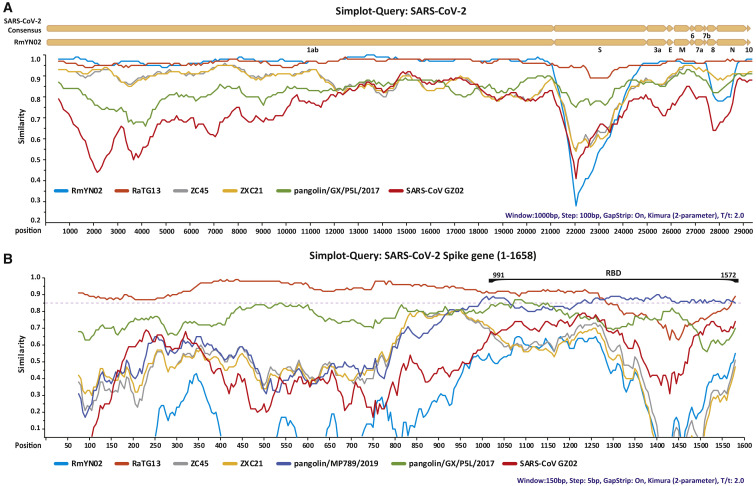
In contrast, RmYN02 was closely related to SARS-CoV-2, exhibiting 93.3% nucleotide sequence identity, although it was less similar to SARS-CoV-2 than RaTG13 (96.1%) across the genome as a whole (Table 1 ). RmYN02 and SARS-CoV-2 were extremely similar (>96% sequence identity) in most genomic regions (e.g., 1ab, 3a, E, 6, 7a, N, and 10) (Table 1). In particular, RmYN02 was 97.2% identical to SARS-CoV-2 in the longest encoding gene region, 1ab (21,285 nucleotides). However, RmYN02 exhibited far lower sequence identity to SARS-CoV-2 in the S gene (nucleotide 71.8%, amino acid 72.9%), compared to 97.4% amino acid identity between RaTG13 and SARS-CoV-2 (Table 1). Strikingly, RmYN02 only possessed 62.4% amino acid identity to SARS-CoV-2 in the RBD, whereas the pangolin beta-CoV from Guangdong had amino acid identity of 97.4% [6], and was the closest relative of SARS-CoV-2 in this region. A similarity plot estimated using Simplot [10] also revealed that RmYN02 was more similar to SARS-CoV-2 than RaTG13 in most genome regions (Figure 1 A). Again, in the RBD, the pangolin/MP789/2019 virus shared the highest sequence identity to SARS-CoV-2 (Figure 1B).
Table 1.
Sequence Identity for SARS-CoV-2 Compared with RmYN02 and Representative Beta-CoV Genomes
| Strain | Complete Genome | Gene Region |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1ab | S | RBD | 3a | E | M | 6 | 7a | 7b | 8 | N | 10 | |||
| Nucleotide sequences | RmYN02 | 93.3%a | 97.2% | 71.9% | 61.3% | 96.4% | 98.7% | 94.8% | 96.8% | 96.2% | 92.4% | 45.8% | 97.3% | 99.1% |
| RaTG13 | 96.1% | 96.5% | 92.9% | 85.3% | 96.3% | 99.6% | 95.4% | 98.4% | 95.6% | 99.2% | 97.0% | 96.9% | 99.1% | |
| ZC45 | 87.6% | 89.0% | 75.1% | 62.1% | 87.8% | 98.7% | 93.4% | 95.2% | 88.8% | 94.7% | 88.5% | 91.1% | 99.1% | |
| ZXC21 | 87.4% | 88.7% | 74.6% | 60.6% | 88.9% | 98.7% | 93.4% | 95.2% | 89.1% | 95.5% | 88.5% | 91.2% | / | |
| pangolin/GD/2019b | – | 90.8% | 89.3% | – | 93.4% | 98.3% | 93.1% | 94.6% | 93.4% | – | 92.1% | 96.1% | – | |
| pangolin/GX/P5L/2017 | 85.2% | 84.7% | 83.2% | 79.9% | 87.0% | 97.4% | 91.3% | 90.9% | 86.6% | 81.8% | 80.6% | 91.0% | 94.0% | |
| SARS-CoV GZ02 | 78.9% | 79.6% | 72.3% | 73.8% | 75.6% | 93.5% | 85.1% | 74.5% | 82.1% | 83.0% | 45.3% | 88.1% | / | |
| Amino acid sequences | RmYN02 | N/A | 98.8% | 72.9% | 62.4% | 96.7% | 100.0% | 98.2% | 96.7% | 95.9% | 83.7% | 27.3% | 98.6% | 97.4% |
| RaTG13 | N/A | 98.5% | 97.4% | 89.3% | 97.8% | 100.0% | 98.6% | 100.0% | 97.5% | 97.7% | 95.0% | 99.0% | 97.4% | |
| ZC45 | N/A | 95.6% | 80.2% | 63.5% | 90.9% | 100.0% | 98.6% | 93.4% | 87.6% | 93.0% | 94.2% | 94.3% | 97.4% | |
| ZXC21 | N/A | 95.2% | 79.6% | 62.9% | 92.0% | 100.0% | 98.6% | 93.4% | 88.4% | 93.0% | 94.2% | 94.3% | / | |
| pangolin/GD/2019b | N/A | 97.1% | 90.7% | 97.4% | 97.4% | 100.0% | 98.6% | 96.6% | 97.5% | – | 94.9% | 97.6% | – | |
| pangolin/GX/P5L/2017 | N/A | 92.6% | 92.4% | 86.8% | 89.8% | 100.0% | 98.2% | 95.1% | 88.4% | 72.1% | 87.6% | 93.8% | 84.2% | |
| SARS-CoV GZ02 | N/A | 86.2% | 76.2% | 74.6% | 73.1% | 94.7% | 89.6% | 68.9% | 85.2% | 79.5% | 29.7% | 90.5% | / | |
Pangolin/GD/2019 and pangolin/GX/P5L/2017 (EPI_ISL_410540). –, no corresponding values in [6]; /, this open reading frame is not found; N/A, not available.
Sequence identities for RmYN02 compared with the SARS-CoV GZ02 (accession number AY390556); the bat SARS-like coronaviruses RaTG13 (EPI_ISL_402131), ZC45 (MG772933), and ZXC21 (MG772934); and the pangolin SARS-like coronaviruses
Pangolin/GD/2019 represents a merger of GD/P1L and GD/P2S, and these values were adapted from [6]
Figure 1.
Patterns of Sequence Identity between the Consensus Sequences of SARS-CoV-2 and Representative Beta-CoVs
(A) Whole-genome similarity plot between SARS-CoV-2 and representative viruses listed in Table 1. The analysis was performed using Simplot, with a window size of 1,000 bp and a step size of 100 bp.
(B) Similarity plot in the spike gene (positions 1–1,658) between SARS-CoV-2 and representative viruses listed in Table 1. The analysis was performed using Simplot, with a window size of 150 bp and a step size of 5 bp.
See also Table S3.
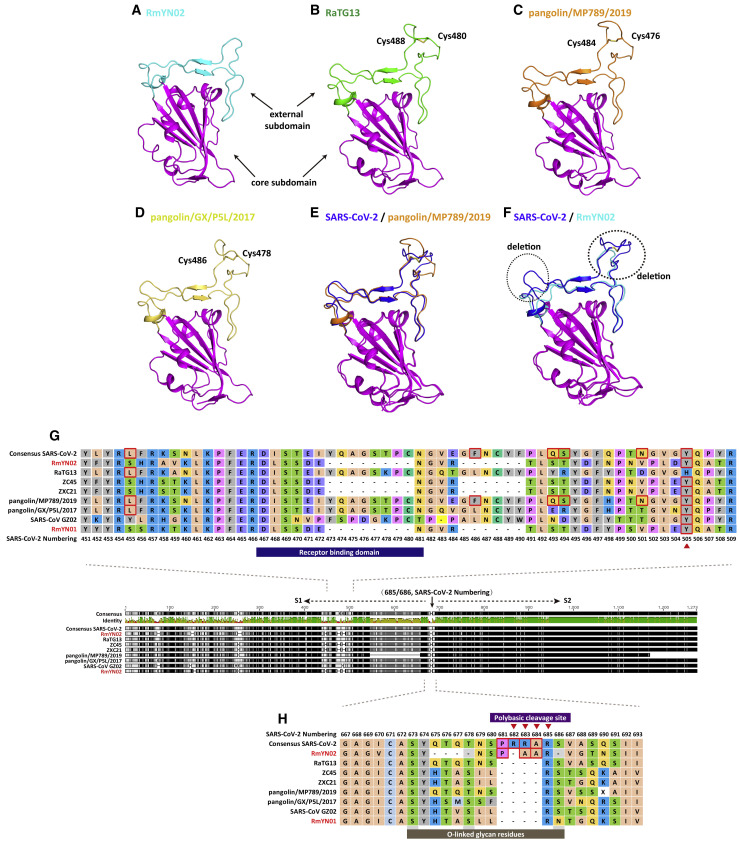
Results from homology modeling [2], in vitro assays [3], and resolved three-dimensional structure of the S protein [11] have revealed that like SARS-CoV, SARS-CoV-2 could also use ACE2 as a cell receptor. We analyzed the RBD of RmYN02, RaTG13, and the two pangolin beta-CoVs using homology modeling (Figures 2 A–2F and S3 for sequence alignment). The amino acid deletions in RmYN02 RBD made two loops near the receptor binding site that are shorter than those in SARS-CoV-2 RBD (Figures 2A and 2F). Importantly, the conserved disulfide bond in the external subdomain of SARS-CoV (PDB: 2DD8) [12], SARS-CoV-2 (PDB: 6LZG), RaTG13 (Figure 2B), pangolin/MP789/2019 (Figure 2C), and pangolin/GX/P5L/2017 (Figure 2D) was missing in RmYN02 (Figure 2F). We speculate that these deletions may cause conformational variations and consequently reduce the binding of RmYN02 RBD with ACE2 or even cause non-binding. It is possible that the bat SARS-related CoVs with loop deletions, including RmYN02, ZXC21, and ZC45, use a currently unknown receptor. In contrast, RaTG13 (Figure 2B), pangolin/MP789/2019 (Figure 2C), and pangolin/P5L/2017 (Figure 2D) did not have the deletions, and had similar conformations at their external domains, indicating that they may also use ACE2 as cell receptor, although with the exception of pangolin/MP789/2019 (see below), all exhibited amino acid variation to SARS-CoV-2. Indeed, the pangolin/MP789/2019 virus showed highly structural homology with SARS-CoV-2 (Figure 2E).
Figure 2.
Homology Modeling of the RBD Structures and Molecular Characterizations of the S1/S2 Cleavage Site of RmYN02 and Representative Beta-CoVs
(A–D) Homology modeling and structural comparison of the RBD structures of RmYN02 and representative beta-CoVs, including (A) RmYN02, (B) RaTG13, (C) pangolin/MP789/2019, and (D) pangolin/GX/P5L/2017. The three-dimensional structures of the RBD from Bat-SL-CoV RmYN02, RaTG13, pangolin/MP789/2019, and pangolin/GX/P5L/2017 were modeled using the Swiss-Model program [13] employing the RBD of SARS-CoV (PDB: 2DD8) as a template. All the core subdomains are colored magenta, and the external subdomains of RmYN02, RaTG13, pangolin/MP789/2019, and pangolin/GX/P5L/2017 are colored cyan, green, orange, and yellow, respectively. The conserved disulfide bond in RaTG13, pangolin/GD, and pangolin/GX is highlighted, while it is missing in RmYN02 due to a sequence deletion.
(E and F) Superimposition of the RBD structure of pangolin/MP789/2019 (E) and RmYN02 (F) with that of SARS-CoV-2. The two deletions located in respective loops in RmYN02 are highlighted using dotted cycles.
(G) Molecular characterizations of the RBD of RmYN02 and the representative beta-CoVs.
(H) Molecular characterizations of the cleavage site of RmYN02 and the representative beta-CoVs.
See also Figures S2 and S3 and Table S2.
Six amino acid residues at the RBD (L455, F486, Q493, S494, N501, and Y505) have been reported to be major determinants of efficient receptor binding of SARS-CoV-2 to ACE2 [14]. As noted above, and consistent with the homology modeling, pangolin/MP789/2019 possessed the identical amino acid residues to SARS-CoV-2 at all six positions [6]. In contrast, RaTG13, RmYN02, and RmYN01 possessed the same amino acid residue as SARS-CoV-2 at only one of the six positions each (RaTG13, L455; RmYN02, Y505; RmYN01, Y505) (Figure 2G), despite RaTG13 being the closest relative in the spike protein. Such an evolutionary pattern is indicative of a complex combination of recombination and natural selection [6, 15].
The S protein of CoVs is functionally cleaved into two subunits, S1 and S2 [16], in a similar manner to the haemagglutinin (HA) protein of avian influenza viruses (AIVs). The insertion of polybasic amino acids at the cleavage site in the HAs of some AIV subtypes is associated with enhanced pathogenicity [17, 18]. Notably, SARS-CoV-2 is characterized by a four-amino-acid insertion at the junction of S1 and S2, not observed in other lineage B beta-CoVs [19][20]. This insertion, which represents a poly-basic (furin) cleavage site, is unique to SARS-CoV-2 and is present in all SARS-CoV-2 sequenced so far. The insertion of three residues, PAA, at the junction of S1 and S2 in RmYN02 (Figure 2H; Figure S2A for results from Sanger sequencing) is therefore of major importance. Although the inserted residues (and hence nucleotides) are not the same as those in RmYN02, and hence are indicative of an independent insertion event, that they are presented in wildlife (bats) strongly suggests that they are of natural origin and have likely been acquired by recombination. As such, these data are strongly suggestive of a natural zoonotic origin of SARS-CoV-2.
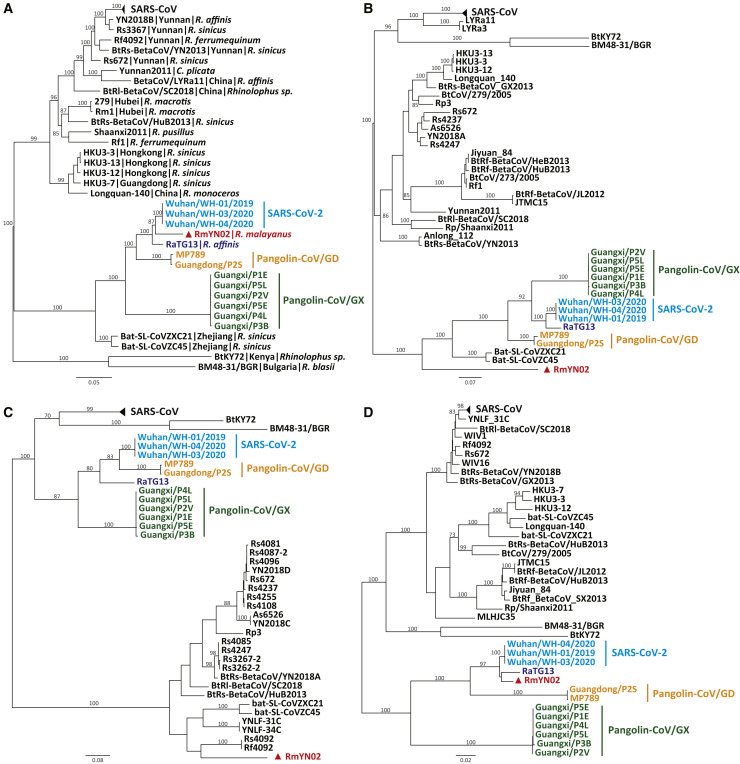
We next performed a phylogenetic analysis of RmYN02, RaTG13, SARS-CoV-2, and the pangolin beta-CoVs. Consistent with previous research [6], the pangolin beta-CoVs formed two well-supported sub-lineages, representing animal seized by anti-smuggling authorities in Guangxi (pangolin-CoV/GX) and Guangdong (pangolin-CoV/GD) provinces (Figures 3 A and S4A). However, whether pangolins are natural reservoirs for these viruses, or they acquired these viruses independently from bats or other wildlife, requires further sampling [6]. More notable was that RmYN02 was the closest relative of SARS-CoV-2 in most of the virus genome, although these two viruses were still separated from each other by a relatively long branch length (Figures 3A and S4A). In the spike gene tree, SARS-CoV-2 clustered with RaTG13 and was distant from RmYN02, suggesting that the latter virus has experienced recombination in this gene (Figures 3B and S4B). In phylogeny of the RBD, SARS-CoV-2 was most closely related to pangolin-CoV/GD, with the bat viruses falling in more divergent positions, again indicative of recombination (Figures 3C and S4C). Finally, phylogenetic analysis of the complete RNA-dependent RNA polymerase (RdRp) gene, which is often used in the phylogenetic analysis of RNA viruses, revealed that RmYN02, RaTG13, and SARS-CoV-2 formed a well-supported sub-cluster distinct from the pangolin viruses (Figures 3D and S4D).
Figure 3.
Phylogenetic Analysis of SARS-CoV-2 and Representative Viruses from the Subgenus Sarbecoronavirus
(A) Phylogenetic tree of the full-length virus genome.
(B) The S gene.
(C) The RBD.
(D) The RdRp.
Phylogenetic analysis was performed using RAxML [21] with 1,000 bootstrap replicates, employing the GTR nucleotide substitution model. RBD is delimited as the gene region 991–1,572 of the spike gene according to [6]. All the trees are midpoint rooted for clarity.
See also Figure S4.
We confirmed the bat host of RmYN02, Rhinolophus malayanus, by analyzing the sequence of the cytochrome b (Cytb) gene from the NGS data; this revealed 100% sequence identity to a Rhinolophus malayanus isolate (GenBank: MK900703). Both Rhinolophus malayanus and Rhinolophus affinis are widely distributed in southwest China and southeast Asia. Generally, they do not migrate over long distances and are highly gregarious such that they are likely to live in the same caves, which might facilitate the exchange of viruses between them and the occurrence of recombination. Notably, RaTG13 was identified from anal swabs and RmYN02 was identified from feces, which is a simple, but feasible way for bats to spread the virus to other animals, especially species that can utilize cave environments.
Our study reaffirms that bats, particularly those of the genus Rhinolophus, are important natural reservoirs for coronaviruses and currently harbor the closest relatives of SARS-CoV-2, although this picture may change with increased wildlife sampling. In this context it is striking that the RmYN02 virus identified here in Rhinolophus malayanus is the closest relative of SARS-CoV-2 in the long 1ab replicase gene, although the virus itself has a complex history of recombination. Finally, the observation that RmYN02 contains an insertion of multiple amino acids at the S1/S2 cleavage site in the spike protein clearly indicates that events of this kind are a natural and expected component of coronavirus evolution [22].
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| BetaCoV/bat/Yunnan/RmYN01/2019 | This manuscript | N/A |
| BetaCoV/bat/Yunnan/RmYN02/2019 | This manuscript | N/A |
| Biological Samples | ||
| Samples are provided in the Table S1 | This manuscript | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| RNAlater Stabilization Solution | Invitrogen | Cat#AM7021 |
| RNAiso Plus reagent | TAKARA | Cat#9109 |
| ReverTra Ace qPCR RT Kit | TOYOBO | Cat#FSQ-101 |
| PerfectStart II Probe qPCR SuperMix | TransGen | Cat#AQ711 |
| TransStart Tip Green qPCR SuperMix | TransGen | Cat#AQ141 |
| Exp Taq DNA Polymerase (Mg2+ and dNTPs plus) | AG | Cat#AG11411 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| Deposited Data | ||
| Raw and analyzed data | This manuscript | NMDC1001304; China National Microbiological Data Center |
| RmYN01 genome | This manuscript | EPI_ISL_412976/NMDC60013004-01; GISAID/China National Microbiological Data Center |
| RmYN02 genome | This manuscript | EPI_ISL_412977/NMDC60013004-02; GISAID/China National Microbiological Data Center |
| partial sequence of spike gene of RmYN02 | This manuscript | NMDCN0000001; China National Microbiological Data Center |
| partial sequence of RdRp gene of RmYN02 | This manuscript | NMDCN0000002; China National Microbiological Data Center |
| partial sequence of cytochrome b (cytb) gene of Rhinolophus malayanus isolate YN-190625 | This manuscript | NMDCN0000003; China National Microbiological Data Center |
| SARS-CoV-2 reference genome sequences from databases are provided in the Table S3 | GenBank / GISAID | N/A |
| Oligonucleotides | ||
| Primer sequences are provided in the Table S2 | This manuscript | N/A |
| Software and Algorithms | ||
| Fastp v0.20.0 | [23] | https://github.com/OpenGene/fastp |
| Bowtie2 v2.3.3.1 | [24] | https://bowtiebio.sourceforge.io/bowtie2 |
| Trinity v2.5.1 | [25] | https://github.com/trinityrnaseq/trinityrnaseq/releases |
| Geneious v11.1.5 | The Biomatters development team | https://www.geneious.com/ |
| PeHaplo | [26] | https://omictools.com/pehaplo-tool |
| MAFFT v7.450 | [27] | https://mafft.cbrc.jp/alignment/software/ |
| RAxML v8.1.6 | [21] | https://cme.h-its.org/exelixis/web/software/raxml/index.html |
| MrBayes v3.2.6 | [28] | http://nbisweden.github.io/MrBayes/ |
| Simplot v3.5.1 | [10] | https://www.mybiosoftware.com/simplot-3-5-1-sequence-similarity-plotting.html |
| SWISS-MODEL | [13] | https://swissmodel.expasy.org/ |
| Other | ||
| Sequencing systems | Illumina | NovaSeq 6000 |
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Weifeng Shi (shiwf@ioz.ac.cn).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Sequence data that support the findings of this study have been deposited in the China National Microbiological Data Center (project accession number NMDC1001304 and sequence accession numbers: NMDC60013004-01, NMDC60013004-02, and NMDCN0000001-NMDCN0000003). The sequences of RmYN01 and RmYN02 generated during this study are also available at the GISAID with accession numbers: EPI_ISL_412976 and EPI_ISL_412977.
Experimental Model and Subject Details
Twenty different species of bats were tested in this study (Table S1). Samples were collected between May and October, 2019 from Mengla County, Yunnan Province in southern China (101.27156323E, 21.91889683N and 21.5932019N, 101.2200914E). Xishuangbanna Tropical Botanical Garden has an ethics committee which provided permission for trapping and bat surveys within this study.
Method Details
Sample collection
Between May and October, 2019, a total of 302 samples from 227 bats were collected from Mengla County, Yunnan Province in southern China (Table S1). These bats belonged to 20 different species, with the majority of samples taken from Rhinolophus malayanus (n = 48, 21.1%), Hipposideros larvatus (n = 41, 18.1%) and Rhinolophus stheno (n = 39, 17.2%). The samples included patagium (n = 219), lung (n = 2) and liver (n = 3), and feces (n = 78). All but three bats were sampled alive and subsequently released. All samples were first stored in RNAlater and then kept at −80°C until use.
Next generation sequencing
Based on the bat species primarily identified according to morphological criteria and confirmed through DNA barcoding, the 224 tissue and 78 fecal samples were merged into 38 and 18 pools, respectively, with each pool containing 1 to 11 samples of the same type (Table S1). Fecal samples were transferred into the RNAiso Plus reagent (TAKARA) for homogenization with steel beads. Total RNA was extracted and subsequently purified using EZNA Total RNA Kit (OMEGA). And tissue samples were extracted using RNeasy Mini Kit (QIAGEN). Libraries were constructed using the NEB Next Ultra RNA Library Prep Kit (NEB). rRNA of feces or tissues was removed using the TransNGS rRNA Depletion (Bacteria) Kit and TransNGS rRNA Depletion (Human/Mouse/Rat) Kit (TransGen), respectively. Paired-end (150 bp) sequencing of each RNA library was performed on the NovaSeq 6000 platform (Illumina) carried out by Novogene Bioinformatics Technology (Beijing, China).
Genome assembly and annotation
Raw reads were obtained from the 56 pools and were then adaptor- and quality- trimmed with the Fastp program [23]. The clean reads were then mapped to reference genomes of representative CoV genomes using Bowtie 2 [24], including SARS-CoV-2 (MDC60013002-01), SARS-CoV (AY508724, AY485277, AY390556 and AY278489), SARS-like-CoV (DQ084200, DQ648857, GQ153542, GQ153547, JX993987, JX993988, KF294455, KF294457, KJ473814, KJ473815, KJ473816, KT444582, KY417142, KY417145, KY417146, KY417148, KY417151, KY417152, KY770859, MK211374, MK211376, and MK211377), ZC45 (MG772933), ZXC21 (MG772934), MERS-CoV (JX869059), other representative beta-CoV genomes (AY391777, EF065505, EF065509, EF065513, FJ647223, KC545386, KF636752, KM349744, KU762338, and MK167038) and alphacoronavirus genomes (NC_002645, AY567487). A total of 11,954 and 64,224 reads in pool no. 39 (a total of 78,477,464 clean reads) were mapped to both a bat coronavirus Cp/Yunnan2011 (JX993988) [9] and SARS-CoV-2, generating two preliminary consensus sequences, named BetaCoV/Rm/Yunnan/YN01/2019 (RmYN01) and BetaCoV/Rm/Yunnan/YN02/2019 (RmYN02), respectively. However, there were only few reads in the remaining 55 pools that could be mapped to these reference CoV genomes. Pool 39 comprised 11 fecal samples from Rhinolophus malayanus collected between May 6 and July 30, 2019.
To validate the two novel CoV genomes, the clean reads of pool 39 were then de novo assembled using Trinity [25] with default settings. The assembled contigs were compared with the consensuses obtained in the previous step and merged using Geneious (version 11.1.5) (https://www.geneious.com). We found that contigs with high and low abundance corresponded to RmYN02 and RmYN01, respectively, with the abundance of RmYN02 5-10 times greater than that of RmYN01. The gaps between contigs of RmYN02 were complemented by re-mapping the reads to the ends of the contigs, which produced the full-length genome sequence of RmYN02. However, due to the limited number of reads available, only a partial genome sequence of RmYN01 was obtained (23395 bp). Reads were then mapped to the full-length genome sequence of RmYN02 using Bowtie 2 to check base consistency at each nucleotide site. Moreover, to perform de novo assembly for pool 39 and to further distinguish RmYN01 and RmYN02, we used PeHaplo [26] with various overlap parameters (70, 80, 100, and 120). The consensus of each run of PeHaplo was then compared, generating the final full-length genome of RmYN02 (29671 bp). The sequence identity between RmYN01 and Cp/Yunnan2011 across the aligned regions was 96.9%, whereas that between RmYN01 and SARS-CoV-2 was only 79.7%.
Bioinformatics analyses
Reference virus genomes were obtained from NCBI/GenBank (https://www.ncbi.nlm.nih.gov/) using Blastn with SARS-CoV-2 as a query. The beta-CoVs from pangolins (Table S3) were retrieved from GISAID (https://www.gisaid.org/). The open reading frames (ORFs) of the verified genome sequences were predicted using Geneious (version 11.1.5). Pairwise sequence identities were also calculated using Geneious. Potential recombination events were investigated using Simplot (version 3.5.1) [10].
The three-dimensional structures of RBD from RmYN02, RaTG13, pangolin/GD and pangolin/GX were modeled using Swiss-Model program [13] using SARS-CoV RBD structure (PDB: 2DD8) [12] as a template.
Multiple sequence alignment of SARS-CoV-2 and the reference sequences was performed using Mafft [27]. Phylogenetic analyses of the complete genome and major encoding regions were performed using RAxML [21] with 1000 bootstrap replicates, employing the GTR nucleotide substitution model (Figure 3). Phylogenetic analysis was also performed using MrBayes [28], employing the GTR nucleotide substitution model (Figure S4). Ten million steps were run, with trees and parameters sampled every 1,000 steps.
Sanger sequencing
Based on the spike gene sequence of RmYN02, a TaqMan-based qPCR was performed to test the feces of pool 39 (Table S2). Pool 39 comprised 11 feces from Rhinolophus malayanus collected between May 6 and July 30, 2019. However, only eight original samples remained after NGS. The results indicated that the fecal sample no. 123 from R. malayanus, collected on June 25, 2019, was positive for RmYN02 (Figure S1). To further confirm the S1/S2 cleavage site and the 1b (RdRp) gene sequence of RmYN02, five pair primers, F1/R1-F4/R4 and F6/R6, were designed for Sanger sequencing (Table S2). The consensus gene sequence of Sanger sequencing of the amplified products was consistent with those from NGS (Figure S2).
Quantification and Statistical Analysis
No statistical analyses were conducted as part of this study.
Acknowledgments
This work was supported by the Academic Promotion Programme of Shandong First Medical University (2019QL006 and 2019PT008), the Strategic Priority Research Programme of the Chinese Academy of Sciences (XDB29010102 and XDA20050202), the Chinese National Natural Science Foundation (32041010 and U1602265), the National Major Project for Control and Prevention of Infectious Disease in China (2017ZX10104001-006), and the High-End Foreign Experts Program of Yunnan Province (Y9YN021B01). W.S. was supported by the Taishan Scholars Programme of Shandong Province (ts201511056). Y.B. is supported by the NSFC Outstanding Young Scholars (31822055) and Youth Innovation Promotion Association of CAS (2017122). E.C.H. is supported by an ARC Australian Laureate Fellowship (FL170100022). A.C.H. was supported by the Chinese National Natural Science Foundation (grant no. U1602265), Mapping Karst Biodiversity in Yunnan, and the the High-End Foreign Experts Program of Yunnan Province (Y9YN021B01, Yunnan Bioacoustic monitoring program). We thank all the scientists, especially Professor Wuchun Cao and Professor Yi Guan, who kindly shared their genomic sequences of the coronaviruses used in this study.
Author Contributions
W.S., Y.B., and A.C.H. designed and supervised research. X.C. and A.C.H. collected the samples. H.Z. and Y.L. processed the samples. T.H. and J.L. performed genome assembly and annotation. H.Z., J.L., and T.H. performed the genome analysis and interpretation. H.S. performed homology modeling. W.S., Y.B., and A.C.H. wrote the paper. X.C., P.W., D.L., J.Y., and E.C.H. assisted in data interpretation and edited the paper.
Declaration of Interests
The authors declare no competing interests.
Published: May 11, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2020.05.023.
Contributor Information
Alice C. Hughes, Email: ach_conservation2@hotmail.com.
Yuhai Bi, Email: beeyh@im.ac.cn.
Weifeng Shi, Email: shiwf@ioz.ac.cn.
Supplemental Information
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S., Wei W., Cheung W.Y., Li W.J., Li L.F. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. Published online March 26, 2020. [DOI] [PubMed] [Google Scholar]
- 7.Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.-J., Li N., Guo Y., Li X., Shen X. Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv. 2020 doi: 10.1101/2020.02.17.951335. [DOI] [Google Scholar]
- 8.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30:1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z., Yang L., Ren X., He G., Zhang J., Yang J., Qian Z., Dong J., Sun L., Zhu Y. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016;10:609–620. doi: 10.1038/ismej.2015.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabakaran P., Gan J., Feng Y., Zhu Z., Choudhry V., Xiao X., Ji X., Dimitrov D.S. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 2006;281:15829–15836. doi: 10.1074/jbc.M600697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020 doi: 10.1101/2020.02.07.939207. [DOI] [Google Scholar]
- 16.He Y., Zhou Y., Liu S., Kou Z., Li W., Farzan M., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 2004;324:773–781. doi: 10.1016/j.bbrc.2004.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monne I., Fusaro A., Nelson M.I., Bonfanti L., Mulatti P., Hughes J., Murcia P.R., Schivo A., Valastro V., Moreno A. Emergence of a highly pathogenic avian influenza virus from a low-pathogenic progenitor. J. Virol. 2014;88:4375–4388. doi: 10.1128/JVI.03181-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F., Bi Y., Wang J., Wong G., Shi W., Hu F., Yang Y., Yang L., Deng X., Jiang S. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J. Infect. 2017;75:71–75. doi: 10.1016/j.jinf.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Duan G., Zhang W., Shi J., Chen J., Chen S. A furin cleavage site was discovered in the S protein of the 2019 novel coronavirus. Chinese Journal of Bioinformatics (In Chinese) 2020;18:1–4. [Google Scholar]
- 21.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B., Zeng L.-P., Yang X.-L., Ge X.-Y., Zhang W., Li B., Xie J.-Z., Shen X.-R., Zhang Y.-Z., Wang N. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13:e1006698. doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen J., Zhao Y., Sun Y. De novo haplotype reconstruction in viral quasispecies using paired-end read guided path finding. Bioinformatics. 2018;34:2927–2935. doi: 10.1093/bioinformatics/bty202. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura T., Yamada K.D., Tomii K., Katoh K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics. 2018;34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the China National Microbiological Data Center (project accession number NMDC1001304 and sequence accession numbers: NMDC60013004-01, NMDC60013004-02, and NMDCN0000001-NMDCN0000003). The sequences of RmYN01 and RmYN02 generated during this study are also available at the GISAID with accession numbers: EPI_ISL_412976 and EPI_ISL_412977.