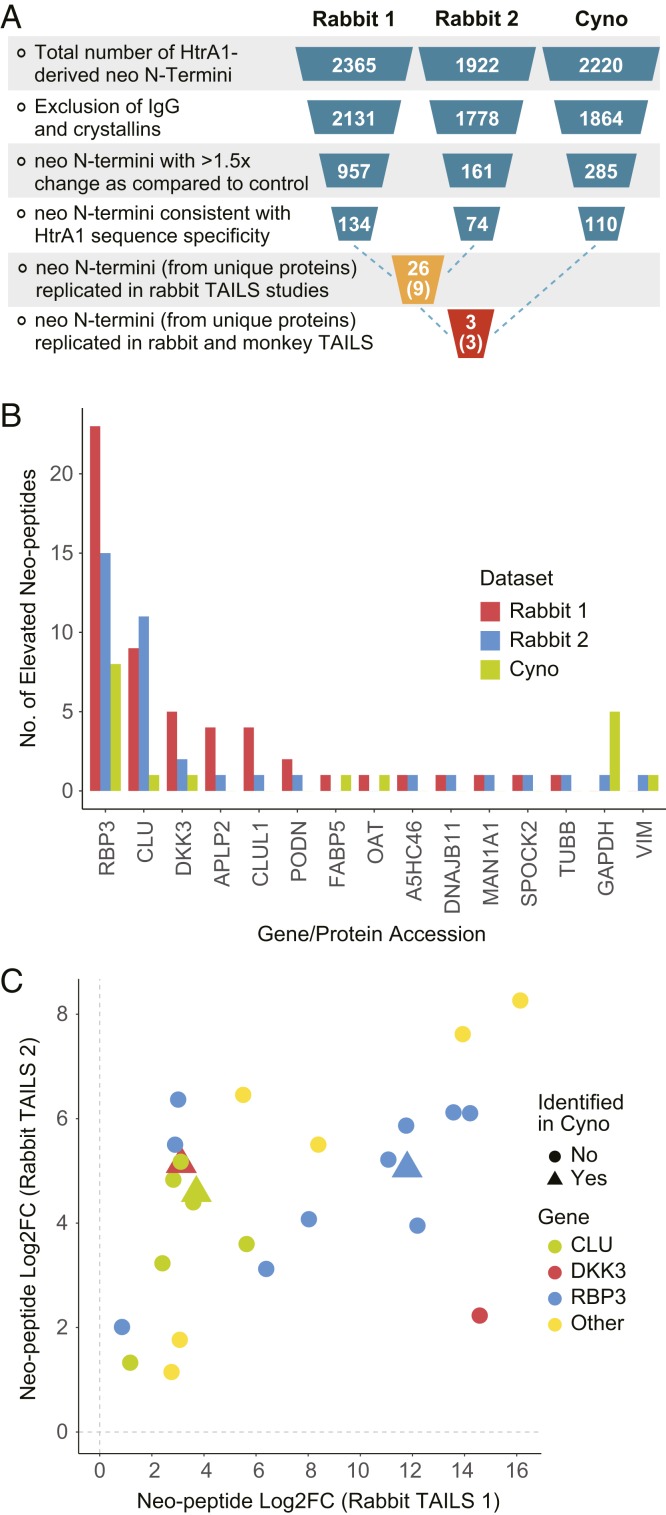
Fig. 5.
Data analysis and triaging of TAILS results. (A) Neo N termini were identified using TAILS and the data were processed as follows: Peptides derived from IgGs and crystallins were excluded and a cutoff of 1.5-fold (log2 fold change [FC] > 0.58) was applied to identify peptides that were enriched in the HtrA1-active as compared with HtrA1-inhibited samples. neo–N-terminal peptides that were not consistent with HtrA1 cleavage-site specificity, for instance, cleavages C-terminal of a basic amino acid, were excluded. From the replicate rabbit studies, we identified 26 peptides with new N termini that were derived from nine proteins. (B) The following proteins were identified based on their neo–N-terminal peptides replicating in the two rabbit studies: RBP3, CLU, DKK3, CLUL1, and APLP2. (C) From the five putative HtrA1 substrates identified in rabbit, only three matching proteins were identified in the TAILS experiment performed in monkey, namely RBP3, CLU, and DKK3. Neo–N-terminal peptides from these three proteins were consistently up-regulated in HtrA1-active as compared with HtrA1-inhibited samples, with similar fold-change magnitude.