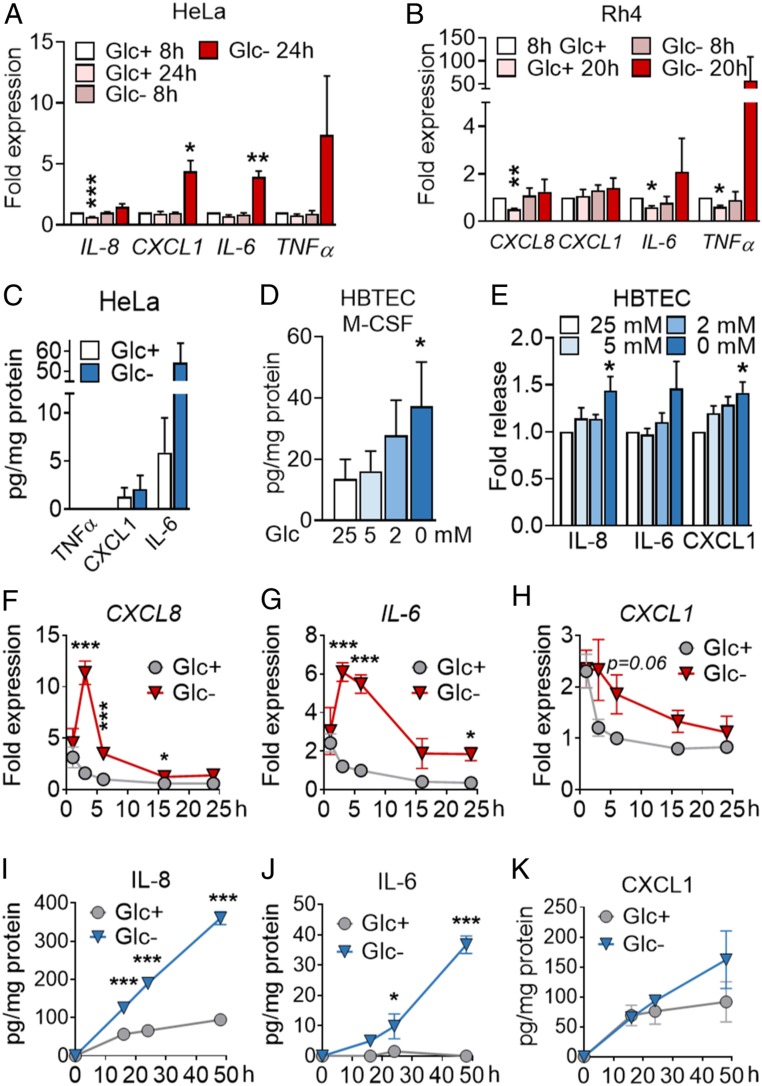
Fig. 1.
Inhibition of glycolysis promotes cytokine synthesis and secretion. (A) HeLa cells were treated for 8 or 24 h with media containing 25 mM (Glc+) or 2 mM (Glc−) glucose. qPCR for indicated genes is shown. Fold expression was calculated by normalizing to control sample Glc+ at 8 h. Data are represented as mean ± SEM (n = 3). Asterisks denote significant differences between treated cells and the 8-h Glc+ sample for each mRNA. (B) Rh4 cells were treated and analyzed for 8 or 20 h as in A. Data represent mean ± SEM (n = 3). Asterisks denote significant differences to the control sample at 8 h in Glc+ of each siRNA. (C) HeLa cells were treated for 24 h with 25 mM or 0 mM Glc, and supernatants were analyzed by ELISA of TNFα, CXCL1, and IL-6. Data represent mean ± SEM (n = 3). (D and E) HBTECs were treated for 24 h with media containing indicated glucose concentrations. Supernatants were analyzed by ELISA for M-CSF (D) and IL-8, IL-6, and CXCL1 (E). Data represent mean ± SEM (n = 3). Asterisks represent significant differences between the 25-mM control sample and each treatment analyzed by one-tailed paired t test. (F–H) A549 cells were treated for 1, 3, 6, 16, and 24 h with media containing 25 (Glc+) or 0 mM (Glc−) glucose. qPCR of CXCL8 (F), IL-6 (G), CXCL1 (H) is shown. Fold expression was calculated by normalizing to cells treated for 6 h with Glc+. Data are represented as mean ± SEM (n = 3–9). Asterisks denote significant differences versus the samples in Glc+ for each time point analyzed by two-way ANOVA. (I–K) A549 cells were treated as in F for indicated time points. ELISA of IL-8 (I), IL-6 (J), and CXCL1 (K) is shown. Data are represented as mean ± SEM (n = 3–4). Asterisks denote significance between the Glc+ and Glc− sample for each time point analyzed by two-way ANOVA. Error bars represent the SEM. The significance was indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001.