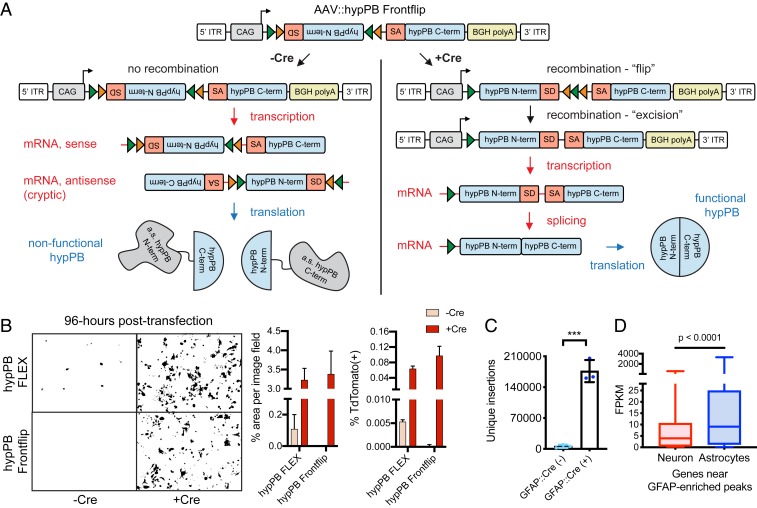
Fig. 6.
AAV::hypPB Frontflip reduces Cre-independent transposition in vitro and in vivo. (A) Schematic of AAV::hypPB Frontflip. Prior to Cre-mediated recombination, hypPB is split into its N and C termini, with the N terminus in reverse orientation and inside a FLEX cassette. On the 3′ end of the N terminus is the splice donor (SD) and 5′ end of an artificial intron. On the 5′ end of the C terminus is the splice acceptor (SA) and 3′ end of the intron. Upon recombination, the N-terminal fragment is flipped into frame, and the artificial intron is reconstituted. This sequence is then transcribed into mRNA, spliced, and translated into an uninterrupted, functional hypPB protein. ITR, inverted-terminal repeat. (B) hypPB FLEX or hypPB Frontflip plasmids were cotransfected into HEK293T cells along with the BrokenHeart fluorescent transposition reporter. At 96-h posttransfection, Cre(−) background is observed from hypPB FLEX but not hypPB Frontflip. For quantification of fluorescent imaging (left graph): n = 2 wells per condition, 3 images per well. For flow cytometry (right graph): n = 2 wells per condition. (C) AAV::hypPB Frontflip significantly reduced transposition in Cre(−) animals (<10,000 insertions per brain), with a >30-fold increase in insertion total in Cre(+) animals (two-tailed, unpaired Student’s t test: ***P < 0.001). GFAP::Cre(+) vs. (−): P = 0.00031, t = 11.63, degrees of freedom = 4, 95% CIdifference = 129,913 to 211,436. (D) Quantifications of neuron- and astrocyte-specific expression of genes near GFAP::Cre-enriched [Neuronmedian = 3.96 FPKM, Astrocytemedian = 9.10 FPKM, n = 615 genes] insertion peaks, called via AAV::hypPB Frontflip data (P < 10−7), showing significant preferential expression in astrocytes (two-tailed Mann–Whitney U test: P < 0.0001).