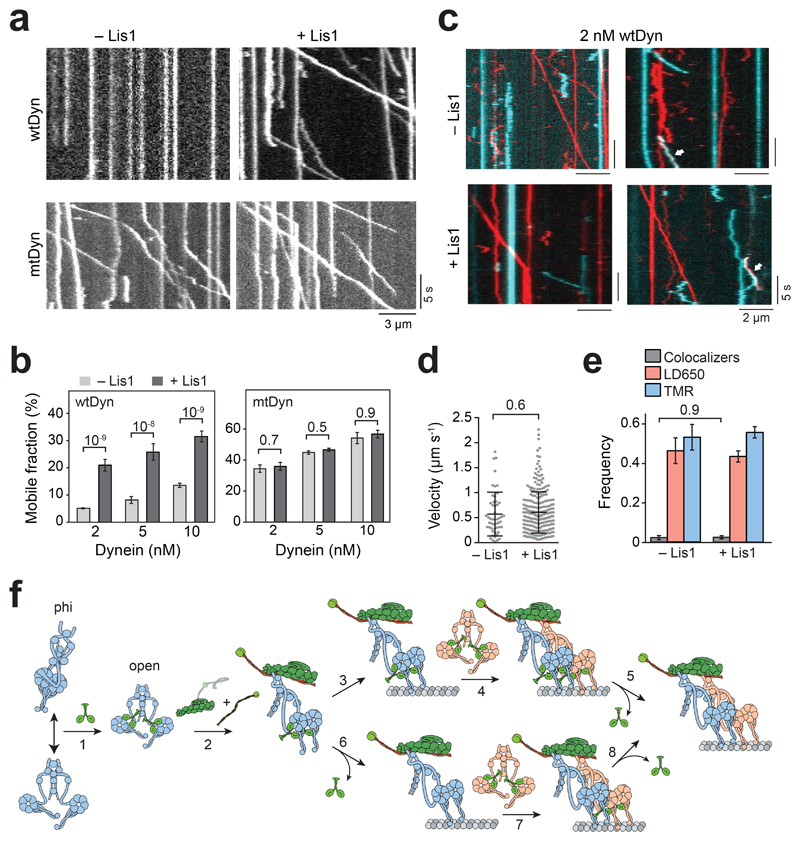
Figure 6. Lis1 promotes assembly of the dynein transport machinery.
(a) Representative kymographs show the motility of DDB at 5 nM concentration of dynein in the absence and presence of 600 nM Lis1. (b) Ratio comparison of the number of processive runs by DDB to the total number of landed motors on MT (mean ± SEM). From left to right, n = 508, 355, 491, 262, 1244, and 392 for wtDyn, and 234, 459, 426, 1352, 457, and 859 for mtDyn. In a and b, three independent experiments were performed per condition. Error bars represent SE calculated from multinomial distribution and p values are calculated from a two-tailed z-test. (c) Representative kymographs show the motility of LD650- (red) and TMR- (cyan) wtDDB at 2 nM concentration of dynein in the absence and presence of 600 nM Lis1. Left kymographs show single-colored runs and right kymographs show rare events of TMR-LD650 colocalization (white arrows). (d) Velocity distribution of wtDDB motility assembled at 5 nM dynein concentration in the absence and presence of 600 nM Lis1. The centerline and whiskers represent the mean and SD, respectively. From left to right, n = 51 and 257, and mean values are 572 and 604 nm s-1. In c and d, three independent experiments were performed per condition. (e) Fraction of processive complexes that contain TMR, LD650, and TMR-LD650 colocalizers (mean ± SEM, n = 59 for -Lis1 and 303 for + Lis1). The p values are calculated from a two-tailed t-test in d and a two-tailed z test in e. (f) A model for Lis1-mediated assembly of the dynein-dynactin complex. (1) Lis1 binds to the open-conformation of dynein with one Lis1 dimer for each dynein motor domain. (2) Lis1 binding prevents transitions of the open conformation to the phi conformation, which increases the affinity of dynein to dynactin. This mechanism also favors the recruitment of second dynein to the complex resulting in higher force production and faster movement. Lis1 dissociates from active dynein-dynactin-cargo adaptor motors, either after pairing of two dyneins with dynactin (3-5) or during the assembly of the complex (6-8).