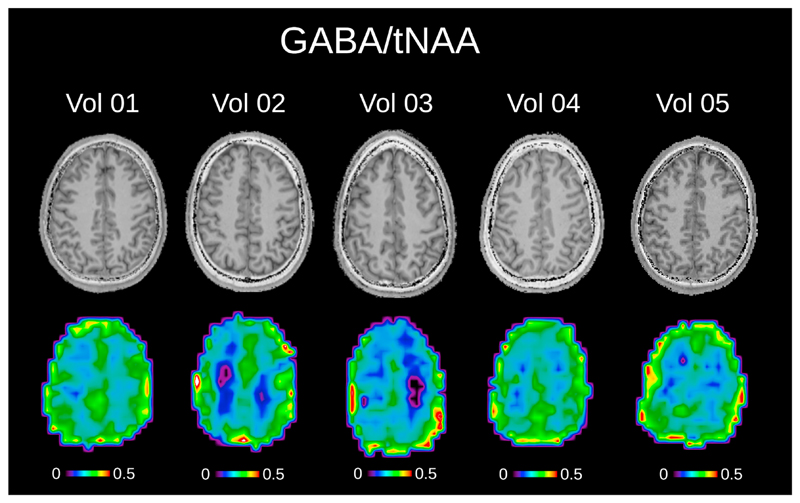
Fig. 7.
The pure GABA/tNAA metabolic maps without MM contamination, as well as the corresponding T1-weighted images are shown for all five volunteers. The tNAA map from EDIT-OFF IR-ON spectra was used as normalization in all cases. Metabolic maps were acquired using the following MRSI settings: FOV 220 × 220 mm2; 32 × 32 matrix; slice thickness 16 mm; TA 24:12 min. The maps were interpolated to a 64 × 64 matrix for display.