Abstract
Glioblastoma multiforme (GBM) is one of the most aggressive types of brain cancer, characterized by rapid progression, resistance to treatments, and low survival rates; the development of a targeted treatment for this disease is still today an unattained objective. Among the different strategies developed in the latest few years for the targeted delivery of nanotherapeutics, homotypic membrane-membrane recognition is one of the most promising and efficient. In this work, we present an innovative drug-loaded nanocarrier with improved targeting properties based on the homotypic recognition of GBM cells. The developed nanoplatform consists of boron nitride nanotubes (BNNTs) loaded with doxorubicin (Dox) and coated with cell membranes (CM) extracted from GBM cells (Dox-CM-BNNTs). We demonstrated as Dox-CM-BNNTs are able to specifically target and kill GBM cells in vitro, leaving unaffected healthy brain cells, upon successful crossing an in vitro blood-brain barrier model. The excellent targeting performances of the nanoplatform can be ascribed to the protein component of the membrane coating, and proteomic analysis of differently expressed membrane proteins present on the CM of GBM cells and of healthy astrocytes allowed the identification of potential candidates involved in the process of homotypic cancer cell recognition.
Keywords: Boron nitride nanotubes, Cell-membrane coating, Homotypic targeting, Glioblastoma multiforme
1. Introduction
The extremely high mortality rate that characterizes glioblastoma multiforme (GBM) makes this tumor one of the deadliest types of cancer, and thus a great challenge for oncologists and biomedical scientists. Although surgery leads to an acceptable alleviation of the symptoms and to a relatively increased life expectancy, the high aggressiveness, the almost universal recurrence, the complex levels of heterogeneity, and the diffuse infiltration tendency of GBM make the treatment of this disease extremely complicated, being thus still characterized by an extremely poor prognosis [1].
Chemotherapy and/or radiotherapy are also used as adjuvant therapies. In this context, doxorubicin (Dox) represents an effective chemotherapy agent, which, however, requires a precise delivery to the tumor site in order to contain its significant side-effects (gastrointestinal toxicity, myelosuppression, cardiotoxicity, and neurotoxicity) [2–4]. Dox has been shown to be toxic on glioblastoma cell lines in vitro, and indeed good performances have been also obtained on in vivo models [5–8], although an efficient targeting system is still required [9].
In recent years, medical research has focused on drug delivery systems and targeted solutions in order to improve drug bioavailability, specifically in proximity of the tumor mass in order to reduce drug toxicity profile [10,11]. The systemic administration of anti-cancer drugs suffers not only from the lack of selectivity, but also from scarce bioavailability, low tumor penetration, limited effectiveness, and from the necessity of repeated administrations with consequent remarkable side effects. The main limitation for the treatment of glioblastoma using anticancer drugs lies on the inability of the majority of these compounds to cross the blood-brain barrier (BBB), which is responsible for the selective transport of small molecules like glucose and amino acids, as well as of small lipid-based substances, from the lumen of the blood capillaries to the extracellular matrix of the central nervous system [12,13]. Nanoparticle-based drug delivery systems, such as, for example, carbon nanotubes, liposomes, or polymeric nanoparticles, offer several advantages with respect to the conventional administration strategies, including an increment of the BBB crossing rate [14], an improved drug targeting/accumulation at tumor level, an enhanced anticancer efficacy, and a lower toxicity in healthy tissues [15–17].
An emerging biocompatible nanoplatform for drug delivery is represented by boron nitride nanotubes (BNNTs), structural analogs of carbon nanotubes. Due to their excellent mechanical and electric properties, and to their chemical stability, BNNTs have been suggested to be used in nanocomposites for the development of advanced functional materials and as novel piezoelectric materials. BNNTs, compared to carbon nanotubes, show a more favorable biocompatibility profile, and based on the current evidence, they can be considered non-toxic [18–20]. Although BNNTs find applications in many fields, their use in the biomedical domain was firstly limited due to their poor solubility in aqueous solvents, because of their high hydrophobicity. Nevertheless, a few attempts based on non-covalent coatings of BNNTs using synthetic or natural polymers demonstrated their potential use in nanomedicine. BNNTs are usually coated with amphiphilic polymers to impart stability in water [21]; their hydrophobicity can be however used to load hydrophobic drugs in their hollow cavity or on their surface [22]: in a recent work, they have been used for example to encapsulate Dox for anti-cancer treatment [23].
Many different ligands have been exploited to promote the targeting of the nanovectors to glioblastoma cells. Molecular “Trojan horses”, such as folic acid, angiopep-2 peptide, and antibodies against transferrin receptors, represent efficient glioblastoma-targeting ligands able to target the nanovectors with their cargo to the cancer site [24–26]. However, the molecular mechanisms involved in cancer recognition are complex phenomena requiring the multiple binding of different ligands to obtain a satisfactory targeting level. Moreover, considering the remarkable genetic heterogeneity of glioblastoma multiforme, the targeting efficacy of the single ligands in different patients is expected to be extremely variable. In this regard, an emerging biomimetic strategy for targeting purposes is the homotypic recognition of tumor cells. This approach exploits the homotypic affinity between cancer cells, mediated by specific membrane proteins, and it relies on the natural properties of cancer cells in developing strong contacts and adhesive interactions [27–29]. Homotypic targeting is achieved by coating or preparing nanoparticles with native extract and components of tumor cell membranes, allowing for the self-recognition with cancer cells [28,30]. This approach also improves the immune tolerance and the nanomaterial stability in biological fluids, such as the blood; therefore, besides being an efficient targeting tool, it also plays a remarkable role for delivery purposes [31–33]. However, the phenomena involved in the homotypic recognition are not completely elucidated, and the comprehension of these molecular mechanisms may enable the developing of a new generation of nanovectors enriched on their surface with a combination of most effective ligands, in order to improve the targeting efficacy of the nanotherapeutics.
In this work, we designed and prepared an innovative nanoplatform consisting of Dox-loaded BNNTs coated with GBM cell membrane (Dox-CM-BNNTs). We demonstrated with multiple in vitro tests that the coating of BNNTs with GBM cell membrane allows selective and efficient targeting of cancer cells. Dox-CM-BNNTs are preferentially internalized by GMB cells, where Dox is released and carries out its pro-apoptotic action. On the other hand, scarce internalization in astrocytes and neural cells has been demonstrated. The role of the protein composition in the nanocarrier targeting has been moreover revealed; finally, the membrane proteins involved in the cancer homotypic recognition have been identified by proteomics and gene ontology analysis.
2. Experimental section
2.1. Cell membrane extraction and BNNT coating
U87 MG cells (ATCC HTB-14™) were maintained in high-glucose (4.5 mg/ml) Dulbecco's modified Eagle's medium (DMEM), supplemented with 1% penicillin-streptomycin (P/S), 1% L-glutamine and 10% fetal bovine serum (FBS). Cells were seeded in 10 cm diameter Petri dishes at a density of 25·103 cells/cm2, and detached with a cell scraper when reached 90% of confluence. After a centrifugation step (750 g for 5 min), pellets were washed twice with Dulbecco's phosphate buffer saline solution (DPBS). After re-suspension in cold (4 °C) ultrapure water, cells were disrupted with a high-pressure homogenizer (20 psi homogenizing pressure). Samples were centrifuged for 10 min at 10000 g, the supernatant containing cell membranes was collected and further centrifuged at 55900 g for 60 min, and the obtained pellet finally re-suspended in 1 ml of de-ionized water.
5 mg of BNNTs (purchased by BNNT LLC) were dispersed in 5 ml of cell membrane extract derived from 25·106 cells; coating was achieved by using an ultrasonic probe (Fisherbrand™ Q125 Sonicator) set at 40 W for 30 min inside an ice-bath. The cell membrane-coated boron ni-tride nanotubes (CM-BNNTs) were collected by centrifugation at 104 g for 15 min at 4 °C, and then washed three times with de-ionized water. The same procedure was used for coating 5 mg of BNNTs with 5 mg of 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)] (mPEG-DSPE), as control sample (mPEG-DSPE-BNNTs). In order to obtain further control samples, cell membranes used for BNNT coating were also pre-treated with two proteolytic enzymes: 0.5 U/mg proteinase K (Sigma Aldrich) for 8 h at 37 °C and, sub-sequently, with trypsin EDTA 0.01% (Thermo Fisher) overnight at 37 °C. After proteolysis, BNNT coating was performed as previously described to obtain de-proteinated cell membrane-coated BNNTs (indicated in the following as CM*-BNNTs). The same procedure was performed to obtain protease-treated mPEG-DSPE-BNNTs, as further control (indicated as mPEG-DSPE*-BNNTs).
2.2. Doxorubicin loading
Dox-loaded BNNTs were obtained by adding 27 μg of Dox (Sigma Aldrich) to 1 mg of BNNTs in 2 ml of ultra-pure water, sonicating for 30 min in ice bath at 20 W, and then incubating for 4 h at room temperature. After incubation, samples were centrifuged for 15 min at 10000 g and washed 2 times to remove non-encapsulated Dox. Cell membrane coating to obtain Dox-CM-BNNTs was performed as described for plain CM-BNNTs. Estimation of Dox loading in Dox-CM-BNNTs was indirectly estimated by assessing the Dox concentrations in the supernatants collected during the washing steps. Dox presence was measured by fluorescence spectroscopy (Agilent Technologies) with an excitation wavelength of 470 nm and an emission wavelength of 590 nm, by exploiting a calibration curve (Fig. S1A).
2.3. Nanovector characterization
The morphological characterization of CM-BNNTs and mPEG-DSPE-BNNTs was performed with bright-field transmission electron microscopy (BF-TEM) and high-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM) imaging, using a FEI Tecnai G2 F20 TWIN TMP with a Schottky emitter operated at 200 kV.
For energy dispersive spectroscopy (EDS) analysis, samples (pristine BNNTs, mPEG-DSPE-BNNTs and CM-BNNTs) have been sonicated for 10 min, and 5 μl of each sample have been deposited on a carbon-coated Cu grid. EDS analyses have been performed using a Bruker XFlash 6|T30 silicon-drift detector (SDD) with 30 mm2 effective area.
Fourier-transformed infrared spectroscopy (FTIR) was performed on freeze-dried samples using a Shimadzu Miracle 10 device. The number of scans was set to 45, the scanning range was set from 4000 to 450 cm−1, and the resolution step at 4 cm−1. The graphs were plotted using Originpro software, 9.1.
X-ray photoelectron spectroscopy (XPS) analyses were performed on a Kratos Axis Ultra DLD spectrometer, using a monochromatized Al Kα source operating at 15 kV and 20 mA. Wide scans were acquired at an analyzer pass energy of 160 eV, while high-resolution narrow scans were performed at constant pass energy of 10 eV and steps of 0.1 eV. The photoelectrons were detected at a take-off angle φ = 0° with respect to the surface normal. The pressure in the analysis chamber was maintained below 7·10−9 Torr for data acquisition. The data were converted to VAMAS format and processed using CasaXPS software, version 2.3.17.
Thermogravimetric analysis (TGA) was performed using a TGA Q50 device from TA Instruments. The temperature was increased from room temperature to 600 °C with a heating rate of 10 °C/min and under a nitrogen flow of 50 ml/min. The graphs were plotted using Originpro software, 9.1.
Dynamic light scattering (DLS) measurements were performed using a Zeta-sizer NanoZS90 (Malvern Instruments LTD). The measurements were carried out at 37 °C in ultrapure water and in cell culture medium (DMEM +10% FBS), and the concentration for all the samples was 100 μg/ml. The Z-potential measurements were carried out in ultrapure water (pH 5.5), and the conductivity was adjusted in the range of 30–100 μS/cm. The hydrodynamic diameter and the Z-potential values represent the mean ± SD of 3 different measurements, with 15 runs for each of them. Before each measurement, the samples were sonicated for 2 min at 30% amplification (Fisherbrand™ Q125 Sonicator) to avoid the presence of aggregates.
Bicinchoninic acid assay (BCA) Protein Kit (Thermo Scientific) was used to determine the amount of protein on the cell membrane-coated surface of the nanovectors, following the manufacturer's protocol. Briefly, 25 μl of 100 μg/ml nanotube dispersions (mPEG-DSPE-BNNTs, CM-BNNT, and CM*-BNNTs) were added in 200 μl of the working solution. After 30 min of incubation at 37 °C, samples were centrifuged for 15 min at 10000 g, and supernatants were collected for absorbance readings at 560 nm (VICTOR X3 plate reader, Perkin Elmer).
2.4. Proteomics investigation of CM proteins through tandem mass spectrometry
Mass spectrometry was performed to identify the proteins of CM-BNNT coating involved in the homotypic targeting of glioblastoma cells. U87 MG and human primary astrocytes from cerebral cortex were grown in 10 cm Petri dishes with high-glucose DMEM supplemented with 10% FBS and 1% P/S. Mass spectrometry analysis was carried out on U87 MG membrane extracts, healthy astrocyte membrane extracts (analogously obtained as described for U87 MG membranes), and coating of CM-BNNTs, since astrocytes represent the healthy cells that degenerate into glioblastoma upon genetic transformation. The protein species were identified in each sample with mass spectroscopy; data-bank consultation was selected grouping proteins present in CM-BNNT coatings and in U87 MG membranes but absent in healthy astrocyte membranes.
CM-BNNT precipitates and cell membrane pellets were treated with 100 μl of 1% sodium deoxycholate (SDC) surfactant in 50 mM ammonium bicarbonate (Ambic), treated in a Thermomixer at 500 rpm for 10 min at 80 °C, sonicated for 5 min on ice (five cycles of 20 s with breaks of 40 s), and finally clarified by centrifugation at 16060 g for 10 min at 5 °C. Only in the case of CM-BNNT sample, a further centrifugation step was required for the complete nanotube removal. The protein concentration in the three sample solutions was determined by BCA, using bovine serum albumin as standard for the calibration curve. Subsequently, protein mixtures were reduced and alkylated using 5 mM dithiothreitol and 20 mM iodoacetamide, respectively, prior to trypsin digestion at 1:100 (w/w) at 37 °C for 16 h. Digested protein samples were then treated with 1% trifluoroacetic acid to remove SDC through acidic precipitation, and supernatants were collected and desalted using Mobicol spin columns, before liquid chromatography tandem mass spectrometry analysis. Peptide solutions were dried under vacuum, dissolved in CH3CN/0.1% HCOOH 2/98 to achieve a final peptide concentration of 0.5 μg/μl, and dedicated to nanoLC-nanoESI-MS/MS analysis on an EASY-nLC coupled to an Orbitrap Fusion mass spectrometer equipped with an EASY-Spray source (ThermoFisher Scientific). 2 μl of each sample were trapped on a PepMap C18 precolumn (2 cm × 75 μm i.d., 3 μm, 100 Å, Thermo Fisher Scientific) and the separation was then achieved in a PepMap C18 column (25 cm × 75 μm i.d., 2 μm, 100 Å, Thermo Fisher Scientific) heated at 50 °C at a flow rate of 300 nl/min. In both cases the mobile phases were CH3CN/0.1% HCOOH 2/98 (phase A) and CH3CN/0.1% HCOOH 98/2 (phase B). The elution program was the following: 0 min, 5% B; 1 min, 5% B; 105 min, 22% B; 120 min, 32% B; 130 min, 90% B; 145 min, 90% B; 146 min, 5% B; 161 min, 5% B. Mass spectra were acquired in positive ion mode, setting the spray voltage at 2.3 kV, the capillary temperature at 275 °C, and the RF lens at 60%. Data were acquired in data-dependent mode with a dynamic exclusion of 60 s; survey MS scans were recorded in the Orbitrap analyzer in the mass range 375–1500 m/z at a nominal resolution of 120000 for 200 m/z in profile mode (AGC target 40000; max inject time 50 ms). Data-dependent tandem MS analyses were performed using a top-speed approach (cycle time of 3 s). Ions were fragmented by HCD (isolation width 1.6 m/z, normalized collision energy 27%) and analyzed in the IT in centroid mode (AGC target 2000, max inject time 300 ms). Monocharged ions did not trigger MS/MS experiments (charge states considered from 2+ to 7+).
To identify peptides, raw data were first converted to .mzML files using MSConvert tool; X!Tandem and OMSSA search tools were used to perform protein identification by querying all the fragment ion spectra against the UniProtKB/Swiss-Prot database (Release August 2018; 20386 sequences) using human taxon as database. A minimum number of three peptides was considered to define an identified protein. False positive hits were filtered out with FDR <1%. Gene ontology (GO) cellular component (CC) analysis was performed using the open source Panther database in order to retrieve proteins involved in cell membrane structure organization and regulation.
Results of GO (CC) analysis were selected by a “bottleneck” approach; a group of proteins identified in both CM-BNNTs and U87 MG cell membranes, but absent in healthy astrocyte membranes, was selected as potential candidates involved in homotypic targeting. A literature inquiry was carried out on this group to attribute the biological role of each candidate, and a subsequent intragroup selection of proteins involved in cell-cell and cell-vesicle interaction was performed.
2.5. Doxorubicin release studies
Dox release from Dox-CM-BNNTs was studied at 4 different experimental conditions: at pH 7.4 to mimic healthy extracellular pH, at pH 7.4 with 100 μM of H2O2 to mimic the intracellular pH in oxidative stress conditions, at pH 4.5 to mimic the acidic organelle environment [23], at pH 4.5 with H2O2 100 μM to mimic the cancer cell environment in oxidative stress conditions. 2 ml of Dox-CM-BNNTs solution (1 mg/ml) was maintained in a shaker at 140 rpm at 37 °C, protected from light. The dispersions were centrifuged (10000 g for 15 min) at different time points (6, 24, 48, 72 and 168 h), and the supernatant was collected for further analysis, whereas the pellet was readily re-dispersed using the same solvents and let under shacking at 37 °C for the following time point measurements. The cumulative amount of the released Dox in the supernatants was evaluated using fluorescence spectroscopy at an excitation wavelength of 470 nm and at an emission wavelength of 590 nm. Fluorescence intensity was then converted into Dox concentration by using calibration curves obtained at the specific pH and H2O2 conditions of the samples (Fig. S1B).
2.6. Nanovector targeting in static conditions
The in vitro targeting ability of mPEG-DSPE-BNNTs, CM-BNNTs, and CM*-BNNTs was evaluated under static conditions using 3 different cell lines: a) neuronal-like cells derived from the SH-SY5Y human neuroblastoma cell line (ATCC CRL-2266™), astrocytes type I clone C8D1A (ATCC CRL-2541™), and U87 MG cells (ATCC HTB-14™), all of them cultured at 37 °C, 5% CO2 and 100% humidity.
8 days before the experiments, 3·103 SH-SY5Y cells/cm2 were seeded on 1.9 cm2 glass coverslips into a 24-well plate with Dulbecco's Modified Eagle's Medium / Nutrient Mixture F-12 (DMEM/F12; Sigma Aldrich) supplemented with 10% of FBS and 1% P/S. At 48 h of culture in proliferative conditions, neural differentiation was induced by replacing the proliferation medium with high glucose DMEM, 1% FBS, 10 μM retinoic acid and 1% P/S, similarly as shown in a previous work [34]; cells were maintained in differentiative conditions for 6 days before performing the targeting experiments. U87 MG and C8D1A cells were seeded (6·103 cells/cm2) on sterilized glass coverslips into a 24-well plate with high-glucose DMEM, 10% FBS, 1% P/S, 1% l-glutamine. Nanotube targeting was tested by incubating 100 μg/ml of the 3 types of BNNT samples in 400 μl of complete medium for 6 and 24 h at 37 °C. After incubation, cells were washed twice with PBS and fixed with 4% paraformaldehyde (PFA; Sigma Aldrich) for 20 min at 4 °C. Cultures were stained with TRITC-phalloidin (1:100; Sigma Aldrich) and Hoechst 33342 (1:1000; Invitrogen) in PBS for 30 min at 37 °C. Confocal microscopy acquisitions were performed using a C2s confocal microscope (Nikon); nanotubes were imaged with 642 nm laser excitation and 650–750 nm laser emission, similarly as described in a previous work [35]. Analysis of the signal area (%) of the cells (TRITC-phalloidin) occupied by BNNTs was performed using NIS Elements software (Nikon).
Quantitative analysis of the efficiency of homotypic targeting was performed through flow cytometry. U87 MG and C8D1A cells were seeded in 24-well plates (6·103 cells/cm2) and cultured for 24 h before the experiment. SH-SY5Y were seeded (6·103 cells/cm2) 8 days before the experiment and induced to differentiation for 6 days before BNNT incubation.
mPEG-DSPE-BNNTs, CM-BNNTs, and CM*-BNNTs were labeled with the fluorescent Vybrant™ DiO Cell-Labeling dye (Invitrogen). 20 μl of DiO was added to 1 mg/ml of nanotube suspension and incubated for 2 h at 37 °C. Samples were thereafter centrifuged (10000 g for 15 min) and washed 3 times, and finally dispersed in phenol red-free complete medium. 100 μg/ml of the labeled mPEG-DSPE-BNNTs, CM-BNNTs, or CM*BNNTs were administered to differentiated SH-SY5Y, C8D1A, and U87 MG cells cultured in 24-well plates. After 24 h of incubation, cells were washed 3 times with PBS, treated with 0.05% Trypsin-EDTA, centrifuged, and suspended in PBS for flow cytometry measurements.
Cytometry was performed with a Cytoflex device (Cytoflex Beckmann). Before analyzing the samples with cells, the fluorescence emissions of 100 μg/ml of mPEG-DSPE-BNNTs, CM-BNNTs, and CM*-BNNTs were analyzed, and the acquisition gain was calibrated in order to detect the same fluorescence intensity from the three types of nanotube preparation. The maximum fluorescence intensity of non-treated control cells was used as threshold to discriminate the BNNT-positive from the BNNT-negative cells.
Inductively-coupled plasma optical emission spectroscopy (ICP-OES) elemental analysis was performed to quantify the amount of boron in the cells, and thus to have further quantitative evaluation of BNNT up-take. SH-SY5Y-derived neurons, C8D1A astrocytes, and U87 MG glioblastoma cells were cultured in T75 flask until reaching 80% of confluence and then incubated for 24 h with nanotubes (mPEG-DSPE-BNNTs, CM-BNNTs, and CM*-BNNTs). After nanotube incubation, cells were washed 5 times with PBS and subsequently detached with 0.05% trypsin-EDTA and centrifuged to obtain pellets ready for analysis.
ICP was carried out by using an iCAP-7600 DUO (Thermo Fisher Scientific). All chemical analyses performed by ICP-OES were affected by a systematic error of about 5%. Samples were dissolved in 800 μl of nitric acid (HNO3 60% v/v) and 200 μl of hydrogen peroxide (H2O2 ≥ 30%) during sonication (65 °C for 2 h). Samples were subsequently diluted using ultrapure water at a final volume of 10 ml; the B concentrations in the different samples were detected at 249.77 nm, and obtained values were normalized for the P concentration as representative of the cell mass (P was detected at 178.28 nm). All measurements have been carried out using a plasma power of 1150 W, a nebulizer gas flow of 0.5 l/min, a cool flow of 12 l/min, and an aux flow of 0.5 l/min. Concentrations of BNNTs were calculated from the concentrations of B by considering a 1:1 B to N ratio.
2.7. Nanovector targeting in dynamic conditions
A home-made fluidic bioreactor was used to investigate the homotypic targeting of CM-BNNTs in dynamic conditions. A scheme of the bioreactor is given in the supporting information (Fig. S2). Briefly, the device was fabricated in poly(methyl methacrylate) (PMMA), and is composed of 3 independent chambers located in 3 channels; each chamber has been designed to host a glass coverslip of 0.9 cm2. The cell cultures (differentiated SH-SY5Y, C8D1A astrocytes, and U87 MG glioblastoma cells) have been seeded and cultured separately on each glass coverslip, and transferred into the fluidic bioreactor on the day of the experiment. More in details, SH-SY5Y cells were seeded (6·103 cell/cm2) 8 days before the experiment on a glass coverslip in proliferative medium (DMEM/F12, 10% FBS, 1% P/S) and, 2 days after seeding, the differentiation was induced by replacing the medium with high-glucose DMEM, 1% FBS, 10 μM retinoic acid and 1% P/S; the cells were maintained in these conditions until the beginning of the experiment. U87 MG and C8D1A were seeded 2 days before the experiment at a density of 6·103 cell/cm2 and cultured with complete medium (high-glucose DMEM, 10% FBS, and 1% P/S). The bioreactor was connected to a peristaltic pump (Ibidi fluidic unit) and perfused with 15 ml of 100 μg/ml BNNT suspensions in high-glucose DMEM with HEPES and 10% FBS at a speed flow of 2 ml/min, for 6 h. During the experiment the bioreactor was placed in an incubator at 37 °C. After 6 h of treatment in dynamic conditions, cells were washed 2 times with PBS, fixed with PFA (4% in PBS for 20 min at 4 °C), and finally stained with TRITC-phalloidin and Hoechst 33342, as previously described. Confocal laser scanning microscopy was performed to evaluate the co-localization signals of BNNTs and cells.
2.8. Cell uptake mechanism investigation
The internalization pathway of CM-BNNTs in U87 MG glioblastoma cells was investigated by confocal fluorescence microscopy.
Pinocytosis was studied by using Cascade Blue hydrazide fluorescent dye (Invitrogen). Briefly, U87 MG cells were seeded at a density of 5·103 cells/cm2 in WillCo dishes and, after 1 day of culture, were incubated for 24 h with 200 μl of phenol red-free HEPES-supplemented complete medium with 300 μM of Cascade Blue hydrazide fluorescent dye and 100 μg/ml of CM-BNNTs. The imaging of nanotubes and pinocytosis vesicles was carried out with a confocal fluorescence microscope (C2 system Nikon), and co-localization analysis was performed by NIS Elements software [36].
Vesicles positive for caveolin-1 and clathrin were detected by immunofluorescence followed by confocal microscopy imaging. 100 μg/ml of CM-BNNTs were administered for 24 h to U87 MG cells previously seeded in WillCo dishes. Afterwards, cells were fixed with 4% PFA for 20 min at 4 °C, washed with PBS, and incubated with a blocking solution containing 10% goat serum (GS; EuroClone) in PBS. Samples were then incubated at 37 °C with anti-caveolin-1 primary antibody or anticlathrin primary antibody (1:150, Abcam) in PBS with 10% GS; after 3 washing steps with 10% GS in PBS, samples were stained with 488 nm Alexa-Fluor secondary antibody (1:100; Millipore), TRITC-phalloidin (1:100; Sigma Aldrich), and Hoechst 33342 (1:1000; Invitrogen). Confocal fluorescence microscopy and co-localization analysis were performed as previously described.
The presence of nanovectors in lysosomes and late endosomes was investigated by using the acidotropic LysoTracker Deep Red dye (Invitrogen). U87 MG cells were seeded in WillCo dishes at a density of 5·103 cells/cm2 and subsequently incubated with phenol red-free HEPES-supplemented DMEM with 10% FBS and 100 μg/ml CM-BNNTs, for 24 and 72 h. Samples were washed twice with PBS and treated with LysoTracker (1:2000) in high-glucose complete medium. After 30 min of staining, cells were washed and stained with Hoechst 33342 (1:1000; Invitrogen) for 15 min at 37 °C. Confocal fluorescence microscopy and co-localization analysis were performed as previously described.
2.9. BBB crossing investigation
The BBB crossing ability of mPEG-DSPE-BNNTs and CM-BNNTs was evaluated under static and dynamic conditions on an in vitro model. Concerning the experiments in static conditions, a commercial system was used (cell culture inserts with 3 μm pores for 24-well plate; Falcon). For testing the BBB crossing in dynamic conditions, a custom designed system with two chambers separated by a 3 μm pore size membrane was exploited. The design and the characterization of the BBB fluidic system are described in a previous paper of our group [37]. Human endothelial cell line hCMEC/D3 were adopted as reliable in vitro model to study nanomaterial crossing [38]. Cells were seeded at the density of 5·104 cells/cm2 on the 3 μm pore size membranes in EndoGRO™-MV-VEGF medium (Millipore) with 5% FBS and 1% P/S. The cells were cultured on the porous membranes for 4 days before performing the investigations on nanomaterial crossing. At day 4, BBB characterization was performed, assessing the trans-endothelial electrical resistance (TEER) by using a volt-Ohm-meter (Millicell® ERS-2 Millipore). The nanovector crossing through the BBB was investigated, as already mentioned, both in static and in dynamic conditions by incubating 100 μg/ml of mPEG-DSPE-BNNTs or of CM-BNNTs in the BBB systems.
Concerning the tests in static conditions, 700 μl of cell medium was placed in the abluminal compartment (i.e., the bottom of the well), while the luminal chamber of the insert was filled with 200 μl of medium containing the nanovectors. The concentration of nanovectors in the abluminal compartment was measured at both 24 h and 72 h with flow cytometry.
The BBB crossing studies in dynamic conditions were carried out by applying to the upper chamber of the fluidic system a flow of 12 ml/min (corresponding to a shear stress of 12 dyne/cm2) with a peristaltic pump (Ibidi fluidic unit system). At 3 h of perfusion, the flow was stopped and the medium of the upper channel was replaced with nanovector-free medium. At 72 h from the nanovector removal, the medium in the abluminal chamber was collected and the nanovector concentration measured by flow cytometry.
Calibration curves showing the numbers of events detected by flow cytometry when testing different amount of nanotubes are reported in Fig. S3.
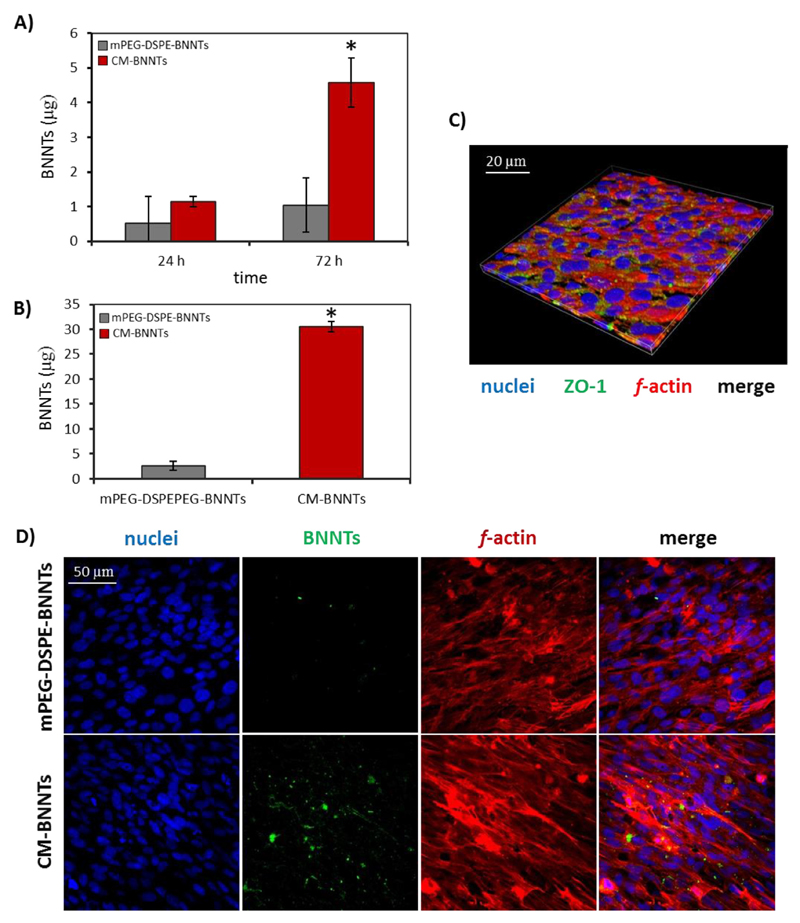
Cells of the models were washed, fixed with 4% PFA (Sigma Aldrich) for 20 min at 4 °C, permeabilized with Triton 0.1% X-100 (Sigma Aldrich) for 15 min, and blocked with 10% GS for 1 h. To evaluate the expression of the tight junctions, the samples were incubated 2 h at 37 °C with 10% GS containing monoclonal anti-tight junction protein 1 (ZO-1) antibody (1:200; Abcam). Samples were thereafter washed in PBS and incubated with Atto 488 goat anti-rabbit secondary antibody (1:500; Sigma Aldrich) in 10% GS at 37 °C for 1 h. Nuclei and f-actin were respectively counterstained with Hoechst 33342 (1:1000; Invitrogen) and TRITC-phalloidin (1:200; Sigma Aldrich). Images were acquired with a confocal microscope (C2s, Nikon).
2.10. Viability assays and therapeutic efficiency
Alamar Blue assay (Invitrogen) was performed, following the manufacturer's protocol, to test viability of U87 MG cells cultured for 1 day in 24-well plates (5·103 cells/cm2) and then treated for 24 and 72 h with 100 μg/ml of mPEG-DSPE-BNNTs, 100 μg/ml of CM-BNNTs, 100 μg/ml of Dox-CM-BNNTs, or with 2.60 μg of free Dox. The amount of the assessed free Dox (2.60 μg) corresponds to the amount of Dox loaded in 100 μg/ml of Dox-CM-BNNT in a final volume of 400 μl of dispersion. After the treatment, cells were washed and incubated with Alamar Blue (1:10 dilution in culture media) for 2 h at 37 °C. The supernatant was then collected, centrifuged, and analyzed with a multi-plate reader (VICTOR X3 Perkin Elmer). Absorbance was measured at both λ1 = 570 nm and λ2 = 595 nm, and then λ1/λ2 ratio was calculated; the viability of cells was expressed as % following normalization with respect to untreated controls.
Immunofluorescence against P53 and Ki-67 markers was performed to investigate apoptosis and proliferation, respectively. P53 protein, as a result of stress factors, enters into the nucleus, activates the expression of pro-apoptotic genes, and inhibits the expression of anti-apoptotic genes [39]. Expression of the Ki-67 antigen in the cell nuclei occurs in phases S, G2, and M of the cell cycle, and in G1 following the mitotic division. The lack of expression of the Ki-67 antigen is observed only in quiescent cells during the G0 phase [40,41]. Staining was performed as described above for clathrin and caveolin-1 markers; after treatment for 24 and 72 h with mPEG-DSPE-BNNTs, CM-BNNTs, or Dox-CM-BNNTs, cells were fixed with 4% PFA in PBS for 20 min at 4 °C. For Ki-67, a primary rabbit anti-Ki-67 antibody (1:150; Abcam), and a FITC-conjugated secondary anti-rabbit antibody (1:150; Millipore) were used; for P53, a primary mouse anti-P53 antibody (1:100; Abcam) and a TRITC-conjugated secondary anti-mouse antibody (1:100; Millipore) were used. Finally, 10 min incubation with Hoechst 33342 in PBS (1:1000; Invitrogen) was performed to stain nuclei. Imaging was performed with a C2s confocal microscope (Nikon); acquisition parameters were calibrated so as to not detect the Dox fluorescence signal. NIS Elements software (Nikon) was used for the analysis of P53 and Ki-67 positive nuclei.
For investigation in fluidic conditions, SH-SY5Y (6·103 cells/cm2) were seeded on glass coverslips and placed in 48-well plates. After 48 h of culture, differentiation was induced for 6 days by using high-glucose DMEM supplemented with 1% FBS, 10 μM retinoic acid, and 1% P/S. C8D1A and U87 MG (6·103 cells/cm2) were instead seeded on glass coverslips in 48-well plates just one day before the experiment. The glass coverslips with the cells were transferred into the previously described bioreactor and exposed to 100 μg/ml of Dox-CM-BNNTs in HEPES-supplemented complete medium for 6 h at 37 °C with a 1.8 ml/min flow speed. After the treatment, the medium containing Dox-CM-BNNTs was replaced with medium without nanotubes, and cells were maintained for 24 h at 37 °C. Finally, cells were fixed, and immunohistochemistry against P53 and Ki-67 followed by confocal fluorescence imaging was performed as previously described.
2.11. Statistical analysis
Once the normality of data distribution was verified with the Shapiro-Wilk test, statistical analysis was carried out with ANOVA followed by Bonferroni post-hoc test.
3. Results and discussion
3.1. Cell membrane coating characterization
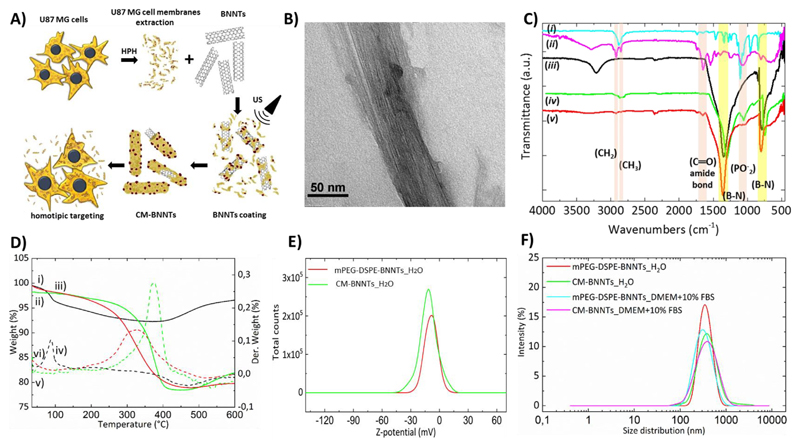
Cell membranes were extracted from the U87 MG cell line, well-characterized and widely proposed in the literature as glioblastoma model [42]. A schematic representation of the cell membrane extraction and the subsequent coating is depicted in Fig. 1A. The coating of BNNTs with deproteinated cell membrane was also performed, aiming at assessing the role of the proteins on the homotypic targeting efficacy. Pictures of the different BNNT preparations, giving a qualitative hint of their stability, are shown in Supporting Information (Fig. S4).
Fig. 1.
A) Schematic representation of the preparation of CM-BNNTs; B) representative TEM image of CM-BNNTs; C) FTIR spectra of mPEG-DSPE (i, cyan curve), CM extract (ii, purple curve), pristine BNNTs (iii, black curve), mPEG-DSPE-BNNTs (iv, green curve), and CM-BNNTs (v, red curve). The characteristic peaks of B—N (yellow), as well as those of C—H, C‗O and P‗O (pink) are highlighted; D) TGA spectra presenting the percentage of weight loss (i, ii and iii) and of its derivative (iv, v and vi) in relation to the temperature increase, for pristine BNNTs (i and iv), mPEG-DSPE-BNNTs (ii and v) and CM-BNNTs (iii and vi); E) Z-potential analysis for mPEG-DSPE-BNNs and CM-BNNTs in ultrapure water; F) size distribution analysis presenting estimated hydrodynamic diameter in ultrapure water and in cell culture medium (DMEM +10% FBS) for mPEG-DSPE-BNNTs and CM-BNNTs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Nanotubes coated with mPEG-DSPE (mPEG-DSPE-BNNTs) were used as control owing to their good stability and water dispersibility, as already described [43,44]. The functionalization procedure relies on a simple non-covalent hydrophobic interaction between lipids and BNNTs, both hydrophobic, that favors the formation of the coating around BNNTs to minimize unfavorable interactions with the aqueous solvent [43]. Lee et al. proposed a possible interaction between mPEG-DSPE and BNNTs where the two fatty acid chains in DSPE are adsorbed or wrapped around BNNTs, while the hydrophilic part, mPEG, allows for the dispersion of functionalized BNNTs in water [43]. Following this description, it is possible to assume that the phospholipids in the cell membrane extract interact in the same way with BNNTs, thanks to their two hydrophobic fatty acid chains, whereas their hydrophilic part (phosphate group) stabilizes the CM-BNNTs in water. Moreover, different studies have postulated interactions between amino acids in the proteins and BNNTs; however, the mechanism and the driving force are not yet fully understood. One possibility is the instauration of hydrophobic interactions between hydrophobic amino acids in proteins and BNNTs. On the other hand, Mukhopadhyay et al. reported, through computational investigation, interactions between BNNTs surface and polar amino acids, like aspartic acid (Asp) and arginine (Arg), through charge transfer and electrostatic interactions, while tryptophan (Trp), a neutral amino acid, had no interaction with the surface of the BNNTs [45]. Another work underlined that the benzene ring in the peptide sequence exhibited a strong π–π interaction with the BNNT surface [46]. In general, Waters et al. have come to the conclusion that BNNTs are able to immobilize proteins through strong interactions with the acidic and basic amino acids [47].
Pristine and coated BNNTs (CM-BNNTs and mPEG-DSPE-BNNTs) were physico-chemically and morphologically characterized. The TEM images in Fig. 1B and Fig. S5A present the morphology of uncoated and coated BNNTs. Despite the cell membrane coating cannot be appreciated with TEM, probably due to its low thickness, several physicochemical characterizations confirm its presence. A first hint was provided by EDS analysis, that showed the co-localization of boron, nitrogen and carbon on the nanotubes (Fig. S5B), suggesting their successful functionalization.
In the FTIR spectra presented in Fig. 1C, it is possible to observe in the traces relative to mPEG-DSPE (i) and CM extract (ii) their typical peaks, in particular related to the stretching of C—H bonds in the –CH2 and –CH3 groups (2750–3000 cm−1) and to the asymmetric vibration between phosphorus and oxygen in the group (1000–1100 cm−1). In the trace (ii) at 1650 cm−1 it also possible to appreciate the typical peak of (C‗O) in the amide bonds, present in the amino acids of cell membrane proteins. Differences between pristine (iii) and coated BNNTs (iv and v) can be observed. In the range of 700–800 and 1250–1400 cm−1, the strong peaks in all the spectra can be attributed to the stretching bonds between boron and nitrogen (B—N). In coated BNNTs, the spectra also show the appearance of the peaks of group in the range 1000–1100 cm−1 (iv and v) and of CH2 and CH3 groups (2750–3000 cm−1) due to the mPEG-DSPE and to the cell membrane extract. In CM-BNNTs (v) it is also possible to observe the peak at 1650 cm−1 of the proteins, as already observed for the plain cell membrane extract (ii). This bond, although not so easily observable, is also present in the mPEG-DSPE-BNNTs spectrum (iv). The peak at approximately 3200 cm−1, that can be observed only in the pristine BNNTs (iii), can be attributed to the hydroxyl groups of water molecules on the BNNT surface. The additional peaks in both mPEG-DSPE-BNNTs and CM-BNNTs spectra (iv and v) support the presence of an external organic coating.
Thermogravimetric analysis was also used as a complementary technique to show the presence of an organic layer around the BNNTs, as well as to calculate the weight percentage of such coating. In the thermograms presented in Fig. 1D, we can observe that the pristine BNNTs start losing weight when the temperature increases above 100 °C, resulting into a maximum weight loss of 7.5%. The initial weight loss can be attributed to the evaporation of water molecules on the surface of the BNNTs, while the increase above 400 °C can be attributed either to the adsorption of nitrogen on their surface (TGA is performed under nitrogen atmosphere) or to impurities that can lead to partial oxidation. TGA analysis finally shows that the coating of both mPEG-DSPE-BNNTs and CM-BNNTs corresponds to approximately 20% of the total weight.
In order to study the stability of pristine and coated BNNTs, we performed dynamic light scattering measurements, evaluating the Z-potential as well as size distribution and polydispersity. The data presented in Fig. 1E present a negative Z-potential of −10.7 ± 0.6 mV and − 19.4 ± 1.2 mV for mPEG-DSPE-BNNTs and CM-BNNTs, respectively. This negative shift of the Z-potential suggests that cell membrane coating may result into higher colloidal stability with respect to mPEG-DSPE. Fig. 1F shows size distribution of coated BNNTs in water and in DMEM +10% FBS. The average hydrodynamic diameter of coated BNNTs (size of 357.5 ± 4.4 nm and PDI of 0.176 ± 0.015 for mPEG-DSPE-BNNTs in water; size of 271.9 ± 45.0 nm and PDI of 0.189 ± 0.035 for mPEG-DSPE-BNNTs in DMEM +10% FBS; size of 366.5 ± 0.6 nm and PDI of 0.208 ± 0.033 for CM-BNNTs in water; size of 332.8 ± 5.5 nm and PDI of 0.213 ± 0.019 for CM-BNNTs in DMEM +10% FBS) indicates a moderate dispersity of the coated BNNTs in water and in complete culture medium (in both cases 0.1 ≤ PDI ≥ 0.4) [48,49]. The slight decrease in hydrodynamic size in DMEM +10% FBS may be due to the presence of serum proteins that further stabilize the nanostructures.
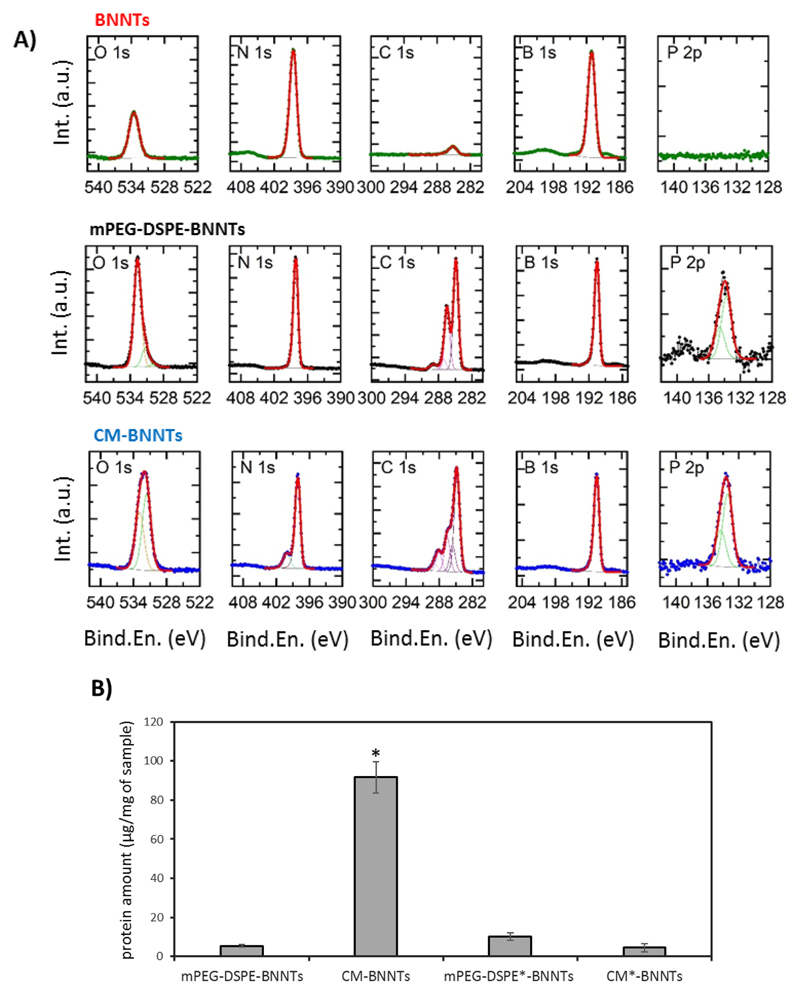
A further demonstration of the presence of CM coating in CM-BNNTs and the quantification of the protein amount is reported in Fig. 2. As expected, the XPS spectra presented in Fig. 2A shows the presence of B 1s and N 1s peaks at respectively 190.6 ± 0.3 eV and 398.2 ± 0.3 eV in all the samples. While the pristine BNNTs present low amount of carbon and oxygen due to environmental contaminations [50], an evident P 2p doublet centered at 133.8 ± 0.3 eV (in agreement with the presence of phospholipids [51]) was found in mPEG-DSPE-BNNTs and in CM-BNNTs, together with stronger contributions in both the C 1s and O 1s regions. Fitting of the high resolution XPS data revealed the presence of C—O, C‗O, and COOH moieties in both mPEG-DSPE-BNNTs and CM-BNNTs samples (as evidenced by the presence of peaks in the carbon spectra at 286.5 ± 0.3, 288.0 ± 0.3, and 289.0 ± 0.3 eV, respectively [52]), suggesting the presence of the organic coating on their surface. Similar results could be obtained by analyzing the O 1s spectra collected on mPEG-DSPE-BNNTs and CM-BNNTs: in both cases, the experimental data fitting revealed the presence of two main components centered at 531.5 ± 0.3 eV and 532.7 ± 0.3 eV, respectively assigned to carboxyl [53] and to ether groups [54] (this second component being the main one in mPEG-DSPE-BNNTs). As a further support to the presence of the cell membrane in CM-BNNTs samples, in the N 1s region, together with the main peak at 398.2 eV assigned to boron nitride [55], a peak at 400.0 ± 0.2 eV is also present, characteristic of uncharged nitrogen, typical of amides and amines [51]. Elements detected by XPS analysis and their % are reported in detail in Table 1.
Fig. 2.
A) XPS analysis of pristine BNNTs, mPEG-DSPE-BNNTs, and CM-BNNTs; B) protein content in mPEG-DSPE-BNNTs, CM-BNNTs, and in the protease-treated mPEG-DSPE*-BNNTs and CM*-BNNTs (* p < .05).
Table 1. XPS elements quantification in pristine BNNTs, mPEG-DSPE-BNNTs, and CM-BNNTs.
| Sample | B (%) | N (%) | C (%) | O (%) | P (%) |
|---|---|---|---|---|---|
| BNNTs | 46.5 | 46.9 | 2.4 | 4.2 | 0.0 |
| mPEG-DSPE-BNNTs | 31.0 | 29.4 | 30.1 | 9.4 | 0.1 |
| CM-BNNTs | 25.2 | 28.5 | 36.4 | 9.7 | 0.2 |
The BCA assay (Fig. 2B) shows the presence of 91.72 ± 8.22 μg of protein in 1 mg of CM-BNNTs. Considering that TGA revealed around 200 μg of cell membrane in 1 mg of CM-BNNTs, we can conclude that proteins represent 45% (w/w) of the coating, a results in line with the average protein content in cell membranes (about 50% w/w) [56]. BCA assay also confirmed the loss of proteins in CM*-BNNTs, that were removed following proteolysis of the cell membrane extracts.
3.2. CM-BNNTs homotypic targeting
The ability of CM-BNNTs to selectively target glioblastoma cells was tested by incubating different cell lines (SH-SY5Y derived neurons, C8D1A, U87 MG) with CM-BNNTs, mPEG-DSPE-BNNTs or CM*-BNNTs.
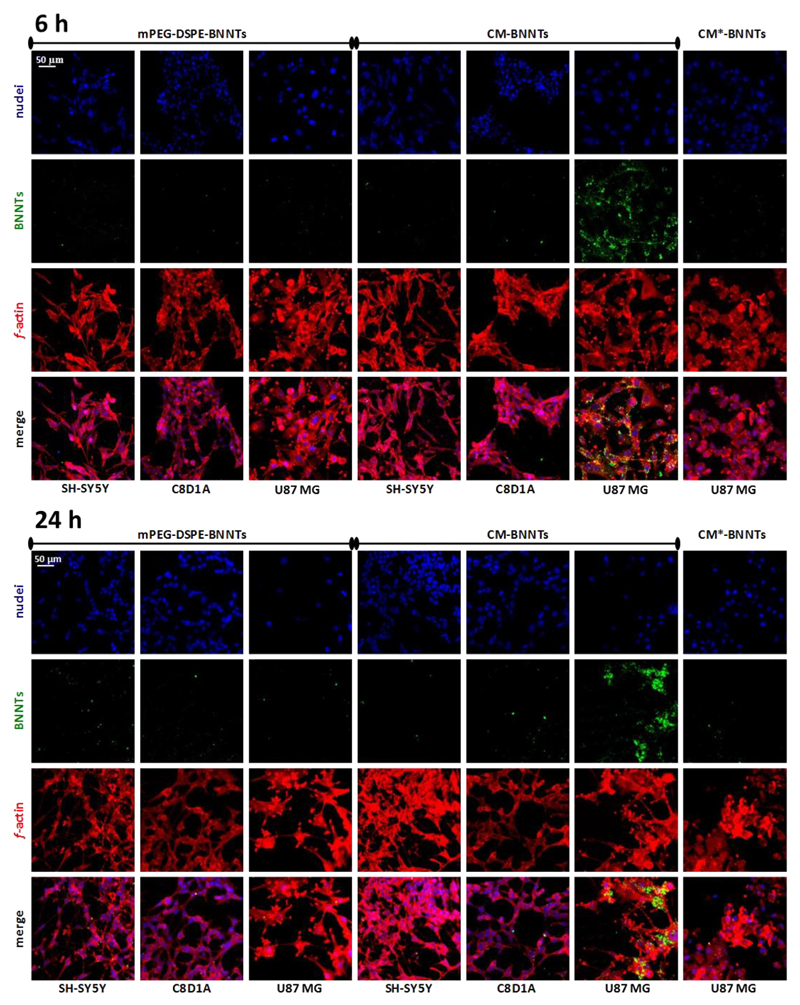
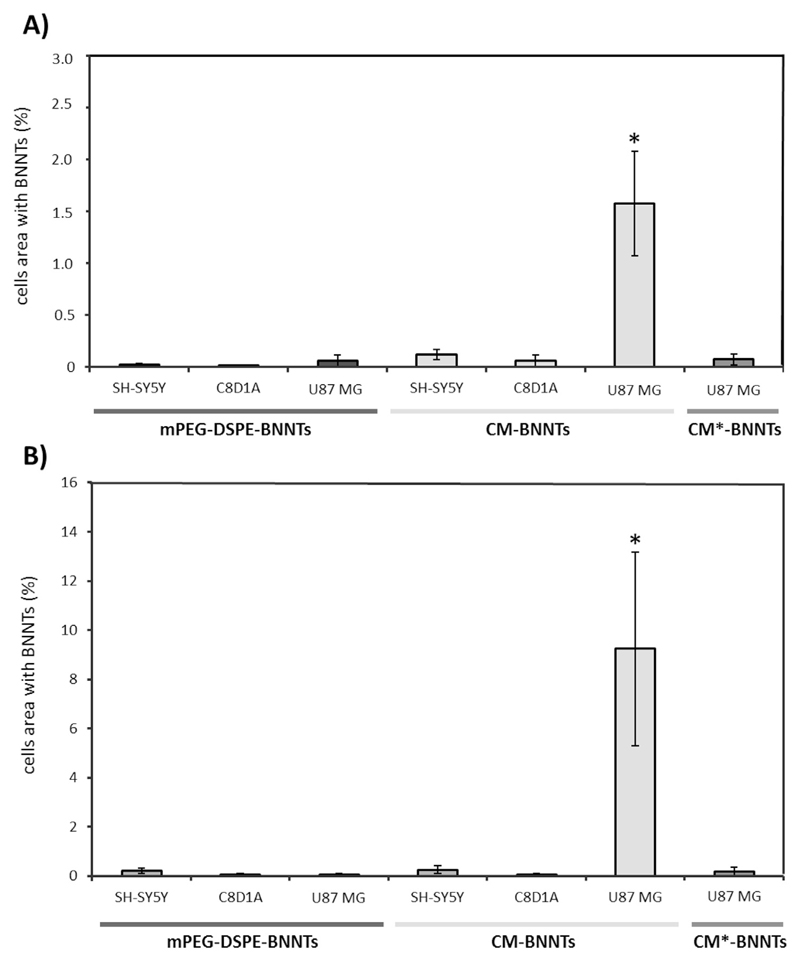
Confocal laser scanning microscopy imaging is shown in Fig. 3, while the respective analysis of the cell area (%) occupied by nanotubes is reported in Fig. 4. Results show an efficient uptake of CM-BNNTs by U87 MG cells. Just after 6 h of treatment, a remarkably higher amount of CM-BNNTs was internalized by U87 MG cells with respect to C8D1A astrocytes (18-folds increase) and to SH-SY5Y derived neurons (11-folds increase). The uptake of CM-BNNTs by U87 MG cells increased after 24 h of incubation, when it resulted 36-fold higher than in SH-SY5Y derived neurons and 154-fold higher with respect to C8D1A astrocytes. Moreover, the uptake of CM-BNNTs in U87 MG cells was 53-fold higher with respect to the uptake of CM*-BNNTs. C8D1A astrocytes and SH-SY5Y derived neurons showed scarce internalization levels for both CM-BNNTs and mPEG-DSPE-BNNTs, suggesting a non-specific interaction with the two BNNT preparations. This cross-incubation experiment demonstrates the relevance of the homotypic targeting for obtaining an efficient and selective nanocarrier delivery to glioblastoma cells. Moreover, the loss of targeting function in CM*-BNNTs revealed the role of the protein component in mediating the membrane-membrane recognition.
Fig. 3.
Targeting investigation. Confocal acquisitions of SH-SY5Y derived neurons, C8D1A astrocytes, and U87 MG cells incubated with 100 μg/ml of mPEG-DSPE-BNNTs, CM-BNNTs, or CM*-BNNTs (f-actin in red, BNNTs in green, nuclei in blue). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Quantitative internalization analysis following confocal microscopy on SH-SY5Y derived neurons, C8D1A astrocytes, and U87 MG cells incubated A) for 6 h and B) for 24 h with 100 μg/ml of mPEG-DSPE-BNNTs, CM-BNNTs, or CM*-BNNTs (* p < .05).
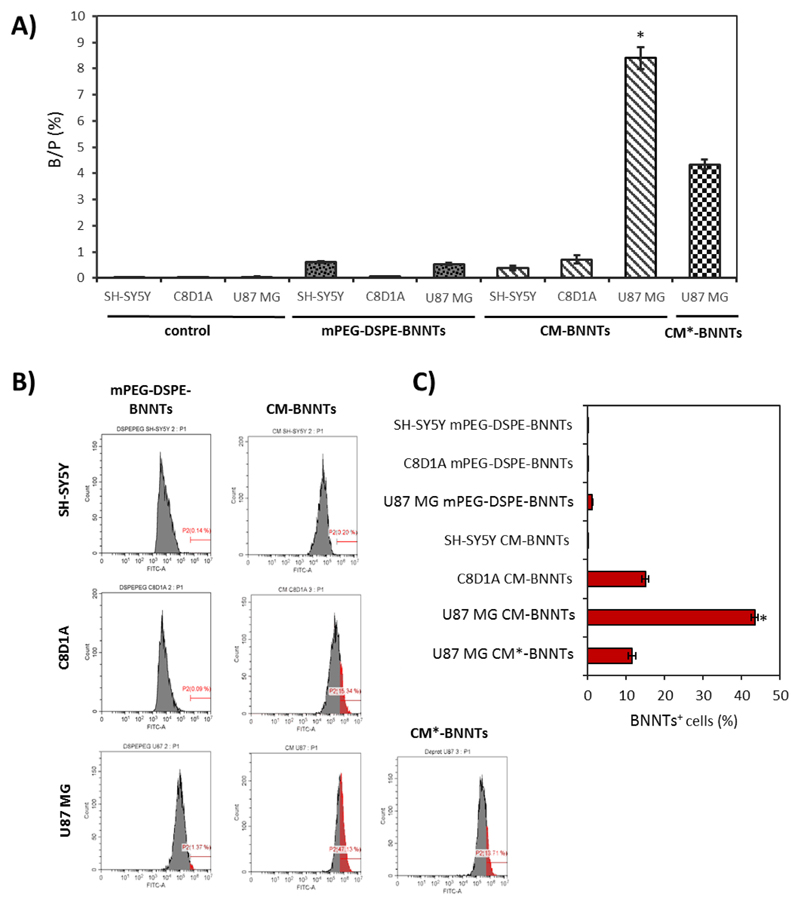
To further demonstrate that a selective homotypic targeting occurs, ICP spectroscopy using boron as target element and flow cytometer analysis were carried out on U87 MG, C8D1A, and differentiated SH-SY5Y incubated for 24 h with nanotubes (CM-BNNTs, mPEG-DSPE-BNNTs, or CM*-BNNTs). ICP analysis in Fig. 5A showed a remarkably higher internalization of CM-BNNTs in U87 MG cells (B/P = 8.39 ± 0.42%) with respect to C8D1A (B/P = .71 ± 0.15%) and to SH-SY5Y derived neurons (B/P = .44 ± 0.08%), while a scarce internalization of mPEG-DSPE-BNNTs was observed in all the cultures (B/P = .53 ± 0.03% in U87 MG cells, B/P = .08 ± 0.01% in C8D1A cells, and B/P = .61 ± 0.03% in SH-SY5Y derived neurons); a remarkably lower uptake of CM*-BNNTs (B/P = 4.33 ± 0.18%) was finally found in U87 MG compared to CM-BNNTs (B/P = 8.39 ± 0.42%), confirming the importance of the protein component of the cell membrane coating in the homotypic targeting.
Fig. 5.
Internalization investigations on SH-SY5Y derived neurons, C8D1A astrocytes, and U87 MG cells treated for 24 h with 100 μg/ml of BNNTs with different coatings. A) ICP investigations on boron content inside cell cultures (* p < .05); B) representative flow cytometry fluorescence data for cells after treatment with DiO-stained BNNTs: for each distribution, populations of BNNT-positive cells are highlighted in red; C) summary of % of BNNT-positive cells for each experimental condition (* p < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similar results were obtained with flow cytometry experiments, where DiO-stained nanotubes were exploited (Fig. 5B and C). The highest percentage of BNNT-positive cells was found in U87 MG cultures treated with CM-BNNTs (43.43 ± 0.83%), while only 11.50 ± 0.99% of U87 MG cells were positive for CM*-BNNTs. A significantly lower amount of CM-BNNT-positive cells was detected in SH-SY5Y (0.19 ± 0.01%) and C8D1A (15.01 ± 0.91%) cultures. Scarce levels of mPEG-DSPE-BNNT internalization were found in all cell cultures (U87 MG 1.21 ± 0.15%, C8D1A 0.09 ± 0.01%, SH-SY5Y 0.09 ± 0.04%).
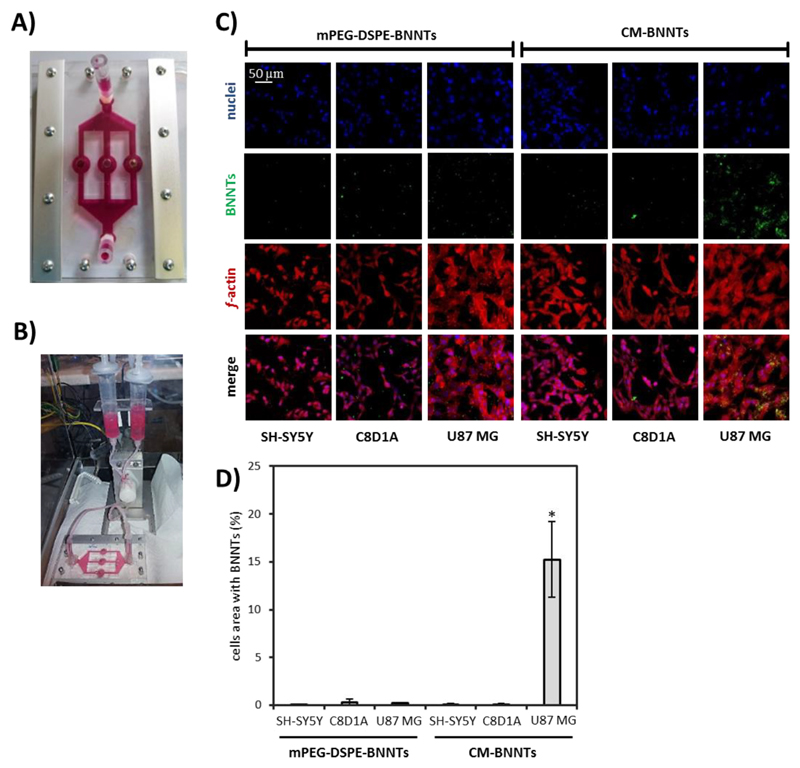
The targeting efficiency of CM-BNNTs was also investigated in dynamic conditions by exploiting a home-made fluidic bioreactor, that allowed to simultaneously test the dynamic targeting of BNNTs in U87 MG cells, C8D1A astrocytes, and SH-SY5Y derived neurons (Fig. 6). The images of this multi-chamber bioreactor during cell culture and during dynamic experiments are shown in Fig. 6A and B, respectively. The confocal fluorescence imaging and the analysis of the nanoparticle internalization are reported in Fig. 6C and D, respectively. Control experiments were carried out by treating cells with mPEG-DSPE-BNNTs. After 6 h of treatment in dynamic conditions (flow speed of 1.8 ml/min for each channel), the internalization of CM-BNNTs in U87 MG cells was 160-fold higher with respect to the other cell cultures incubated with CM-BNNTs, demonstrating a high affinity and selectivity of CM-BNNTs to GBM cells. A scarce internalization of mPEG-DSPE-BNNTs was detected in all the cell cultures. This result suggests as the superior internalization of CM-BNNTs by U87 MG cells is not simply due to a higher uptake rate by this cell line, but, instead, is related to the specific surface properties of the CM coating. Altogether, these evidences proved that CM-BNNTs represent an efficient targeting system, being able to preferentially interact with the GBM cell line thanks to self-recognition properties, thus avoiding extensive accumulation in non-specific healthy cells.
Fig. 6.
CM-BNNTs targeting investigations in dynamic conditions. A) Picture of the multi-compartmental bioreactor used for selective targeting experiments; B) picture of the multi-compartmental bioreactor connected to the fluidic system; C) confocal acquisitions of SH-SY5Y derived neurons, C8D1A astrocytes, and U87 MG cells treated for 6 h with 100 μg/ml of BNNTs in dynamic conditions (f-actin in red, BNNTs in green, nuclei in blue); D) co-localization analysis of BNNTs and cell fluorescence signals (* p < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
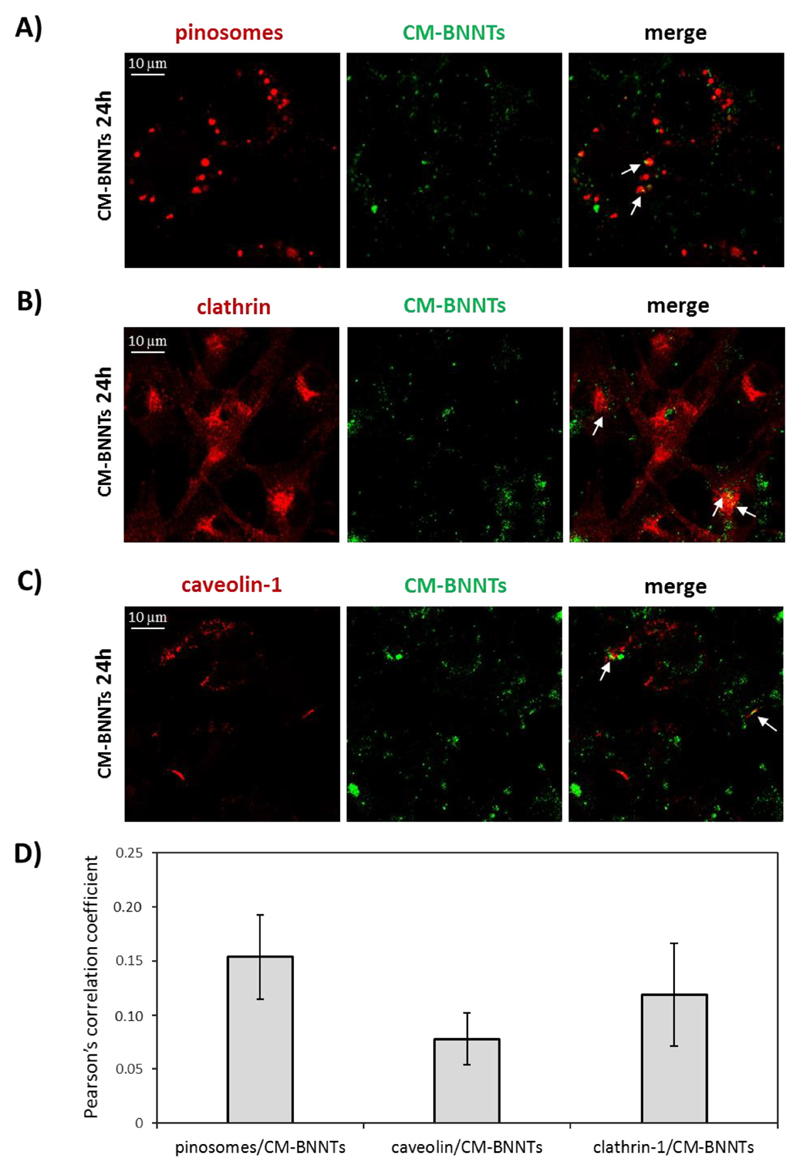
The intracellular pathway of CM-BNNTs in U87 MG cells was investigated through confocal laser scanning microscopy (Fig. 7). The analysis of the CM-BNNT localization in pinosomes, in caveolin-1-positive organelles, and in clathrin-coated vesicles is reported in Fig. 7A, B, and C, respectively. A scarce and similar level of CM-BNNT internalization in all the considered compartments was found. The image analysis reported similar co-localization values (Fig. 7D), indicating that CM-BNNTs are not internalized by a preferential route, but, rather, by multiple internalization pathways. This phenomenon is similar to what observed for multivalent targeting agents, and can be attributed to the multiple and complex protein interactions that occur in homotypic membrane-membrane recognition [57]. However, the investigation of the role of single CM receptors in mediating specific pathways of CM-BNNT internalization is extremely challenging, considering that single CM receptors can be involved in multiple endocytotic pathways [57,58].
Fig. 7.
Analysis of CM-BNNT internalization in U87 MG cells. Confocal fluorescence detection of A) pinosomes, B) caveolin-1, and C) clathrin; D) co-localization analysis of pinosomes/CM-BNNTs, caveolin/CM-BNNTs and clathrin-1/CM-BNNTs fluorescence signals.
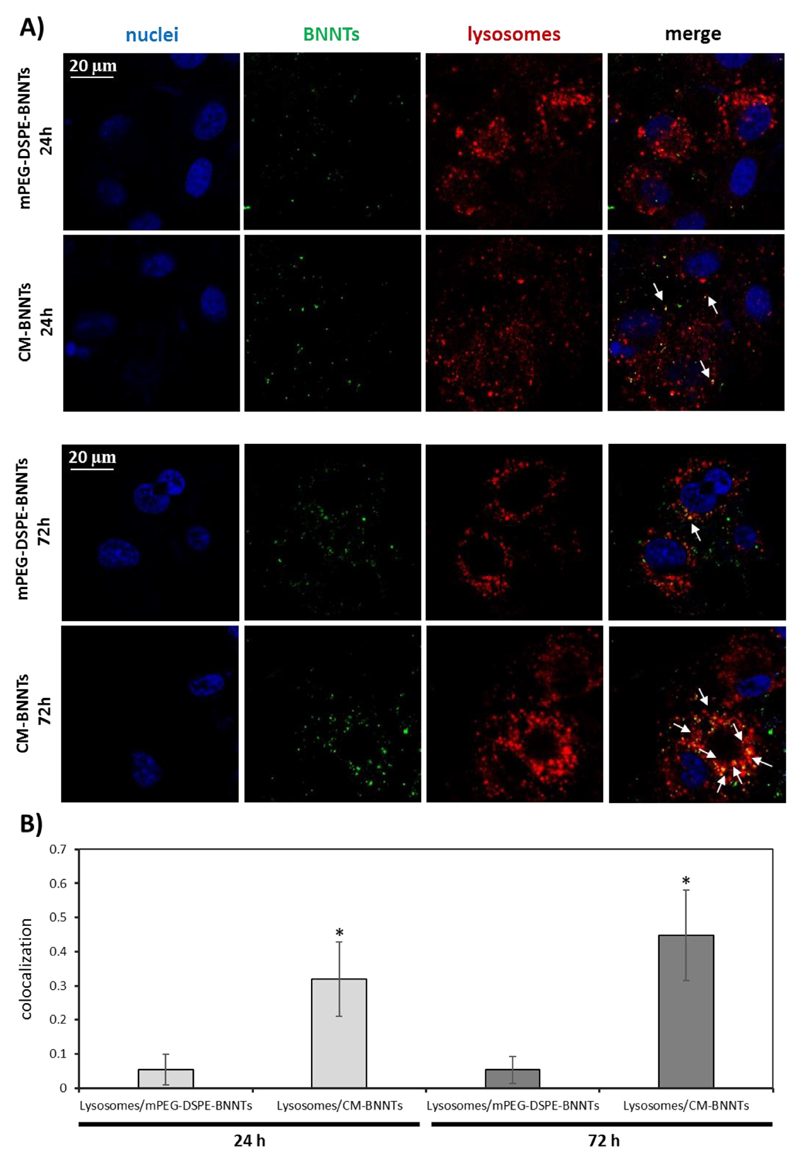
Lysosomal accumulation has also been analyzed (Fig. 8), since this could affect drug release behavior from the nanovectors [59]. Confocal fluorescence imaging of the U87 MG cells incubated for 24 and 72 h with mPEG-DSPE-BNNTs or CM-BNNTs is reported in Fig. 8A. Qualitatively, mPEG-DSPE-BNNTs resulted scarcely localized in acidic compartments, while most of the CM-BNNTs were found in lysosomes at both time points: at 72 h, a significantly higher colocalization between CM-BNNTs and lysosomes (0.448 ± 0.130) was found with respect to mPEG-DSPE-BNNTs (0.053 ± 0.04; Fig. 8B).
Fig. 8.
Analysis of BNNT internalization in acidic organelles (i.e., lysosomes and late endosomes) at 24 and 72 h of incubation with 100 μg/ml of mPEG-DSPE-BNNTs or CM-BNNTs. A) Confocal fluorescence microscopy imaging: lysosomes in red, BNNTs in green, nuclei in blue; B) co-localization analysis between lysosome and BNNT signals (* p < .05). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
A nanovector for brain cancer therapy should be designed to be non-invasive and able to target both the BBB and cancer cells [60]. Specific surface features, charge, shape, size, and functionalization are required for the nanovectors to remain stably suspended in body fluids, to perform targeting, and to cross the BBB [61,62]. BBB crossing experiments were performed in order to test CM-BNNT ability to overcome a BBB-mimicking in vitro endothelial barrier in both static and dynamic conditions. The TEER values (70.05 ± 4.25 Ω cm2) indicated as the BBB models displayed good barrier performances.
Fig. 9A shows the amount of mPEG-DSPE-BNNTs and CM-BNNTs that crossed the in vitro BBB in static conditions after 24 and 72 h of incubation. Scarce amounts of mPEG-DSPE-BNNTs (0.52 ± 0.78 μg) and of CM-BNNTs (1.14 ± 0.15 μg) were detected at 24 h; however, the nanovector crossing significantly increased at 72 h for CM-BNNTs (4.57 ± 0.71 μg, ~4-folds increment), but not in the case of mPEG-DSPE-BNNTs (1.04 ± 0.78 μg).
Fig. 9.
BBB model crossing investigations. mPEG-DSPE-BNNTs and CM-BNNTs crossing the BBB model in A) static and B) dynamic condition (* p < .05); C) immunostaining of the endothelial cells of the BBB model (ZO-1 in green, f-actin in red, nuclei in blue); D) representative confocal images showing BNNT uptake by endothelial cells (f-actin in red, nuclei in blue, BNNTs in green). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Concerning the BBB model in dynamic conditions (Fig. 9B), CM-BNNTs showed again a higher crossing tendency (30.47 ± 47 μg after 72 h) with respect to mPEG-DSPE-BNNTs (2.62 ± 0.92 μg). Representative images of nanovector uptake by endothelial cells are shown in Fig. 9C and D. Collectively, these data demonstrate that the CM coating conferred a double functionality, improving both BBB crossing and cancer cell targeting; this result is in line with recent evidences that showed as component of cancer cell membranes promote transcytosis through the BBB owing to a modulation of the endocytic pathway in brain endothelial cells [63].
Proteomics analysis allowed the identification, with a FDR (false discovery rate) < 1%, of proteins in CM-BNNT coating and in U87 MG/healthy astrocyte membrane extracts; the results (data not shown) indicated as the membrane proteins present in CM-BNNTs are mostly represented by cytoplasm membrane proteins, but also, in minor extent, by endoplasmatic reticulum membrane proteins (8.6% of the total membrane proteins) and by other organelle membrane proteins (4.1% of the total membrane proteins). “Bottleneck” analysis was performed considering only the cell membrane proteins, which were grouped in Venn diagram (Fig. S6). The “bottleneck” analysis individuated 117 proteins that were found both in CM-BNNT coating and in U87 MG membranes, but that were absent in healthy astrocyte membranes; among them, a list of 15 proteins with specific biological function potentially involved in cell-cell recognition was identified (Table 2).
Table 2.
List of cell membrane proteins selected by the “bottleneck” analysis basing on their function. This list includes proteins involved in protein-protein interaction, protein-vesicle interaction, and protein-cytoplasm membrane interactions that were found in CM-BNNT coating and in U87 MG membrane extracts, but not in healthy astrocytes membrane extracts.
| Selected CM-BNNT proteins | Molecular function | Ref. |
|---|---|---|
| Alpha adducin (ADD1) | Promote assembly of spectrin actin network | 71 |
| Cadherin-2 (CDw-325) neural cadherin, (CD antigen CD325) | Cell-cell interaction | 64, 65 |
| CD44 antigen (CDw44) | Cell-cell interaction | 72 |
| N-Cadherin | Cell-cell interaction | 63, 72 |
| Catenin beta-1 (Beta-catenin) | Cell adhesion regulation | 74 |
| Catenin delta-1 (Cadherin-associated Src substrate)(CAS)(p120 catenin) | Cell adhesion regulation | 85 |
| Caveolae-associated protein 3 (Cavin-3) | Regulates caveola formation and trafficking | 75 |
| Neuroplastin (Stromal cell-derived receptor 1) (SDR-1) | Cell-cell interaction | 70 |
| Ras-related protein (Rab-23) | Intracellular membrane trafficking | 76 |
| Synaptosomal-associated protein 23 (SNAP-23) | Modulate membrane shaping and remodeling | 77 |
| Syntaxin-16 (Syn16) (STX16) | Vesicular transport | 78 |
| Tight junction protein ZO-1 (Tight junction protein 1) | Plays a role in tight junction and adherens junction | 79, 80 |
| Tight junction protein ZO-2 (Tight junction protein 2)(Zona occludens protein 2) | Plays a role in tight junction and adherens junction | 81, 82 |
| Tumor cell-derived collagenase stimulatory factor (TCSF)(CD antigen CD147) | Cell-cell interaction | 83 |
| Zyxin (Zyxin-2) | Focal adhesion | 84 |
Among these proteins, cadherin-2 (CDw325) and neuroplastin (stromal cell-derived receptor 1, SDR-1) are well known in the literature for their role in homotypic recognition [64,65]. Cadherins are calcium-dependent cell adhesion proteins that preferentially mediate homotypic cell-cell adhesion by dimerization [66,67]. When the expression of cadherin-2, β-catenin (also found and present in the final list of our targeting proteins), and E-cadherin is altered, dysfunctions connected with tumor invasion and progression may occur [64,65]. In this context, β-catenin and N-cadherin promote the maturation of cell-cell contacts by developing a molecular complex with the actin cytoskeleton, and are considered therapeutic targets and prognostic markers for brainstem glioma [68,69]. Another protein directly involved in homotypic recognition is neuroplastin (SDR-1), that belongs to the superfamily of the cell adhesion molecules (CAMs), and is able to promote the cell adhesion through homophilic interactions [69].
We hypothesize that cadherin-2 and neuroplastin are the main mediators of homotypic targeting of CM-BNNTs toward glioblastoma cells because of their role in binding other proteins of the same family and in developing protein-protein junctions. However, we want to underline that all the proteins listed in Table 2 may cooperatively contribute to the targeting and internalization process, since each of them is involved in cell-cell interactions and in endocytosis processes [68,70–85].
3.3. CM-BNNTs as drug delivery nanovectors
The main aim of this work was to exploit CM-BNNTs to efficiently deliver the anti-cancer drug doxorubicin in GBM cells, without affecting healthy cells. Drug loading measurement highlighted as CM-BNNTs resulted in being efficient nanocarriers, able to load 26.35 μg of Dox per mg of nanotubes (Dox-loaded CM-BNNTs will be now indicated in the text as Dox-CM-BNNTs), corresponding to a drug loading of 2.15% (considering the final weight after the membrane coating) and to an encapsulation efficiency of 95.6%.
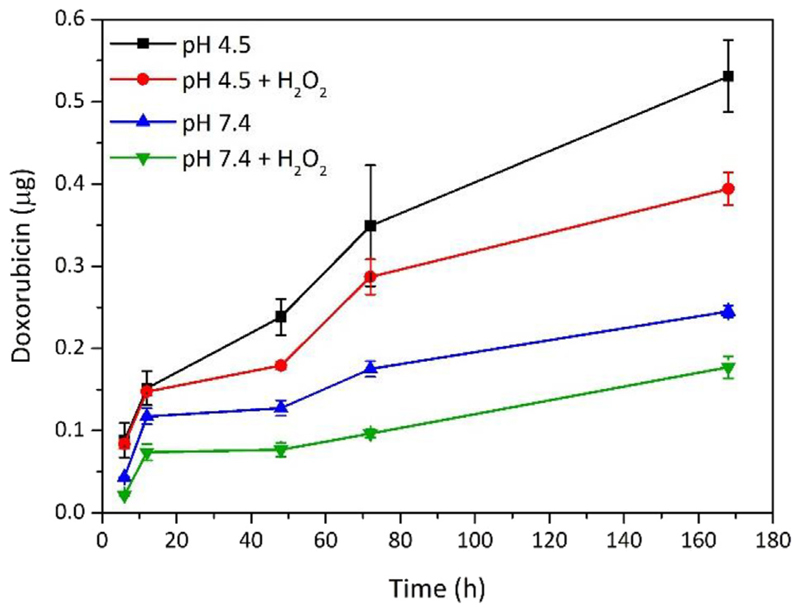
Dox release from Dox-CM-BNNTs was studied at different experimental conditions, as explained in the methods section. The release of the drug from Dox-CM-BNNTs is slow, progressive, and pH-dependent (Fig. 10). The cumulative release at 168 h was higher at pH 4.5 (0.54 ± 0.04 μg of drug, corresponding to 2.0% of total loaded Dox) with respect to pH 7.4 (0.24 ± 0.01 μg of drug, corresponding to 0.75% of total loaded Dox). This is in line with previous studies, where a higher drug release from BNNTs in acidic conditions was found [23,86]. This can be considered an important property of the nanocarriers, which mostly release the drug molecules after their internalization in acidic compartments of cells, thus avoiding the drug release in extracellular environment. Moreover, since solid tumors are characterized by a microenvironment with increased acidity, the preferential drug release from Dox-CM-BNNTs at low pH values can be exploited for a selective drug release at tumor level; this property, combined with the excellent targeting efficiency of the nanocarriers to GBM cancer cells due to the homotypic membrane-membrane recognition, is a hint of an excellent anti-cancer nanoplatform [87].
Fig. 10.
Doxorubicin release from Dox-CM-BNNTs at different conditions (pH 4.5, pH 4.5 + H2O2, pH 7.4, pH 7.4+ H2O2).
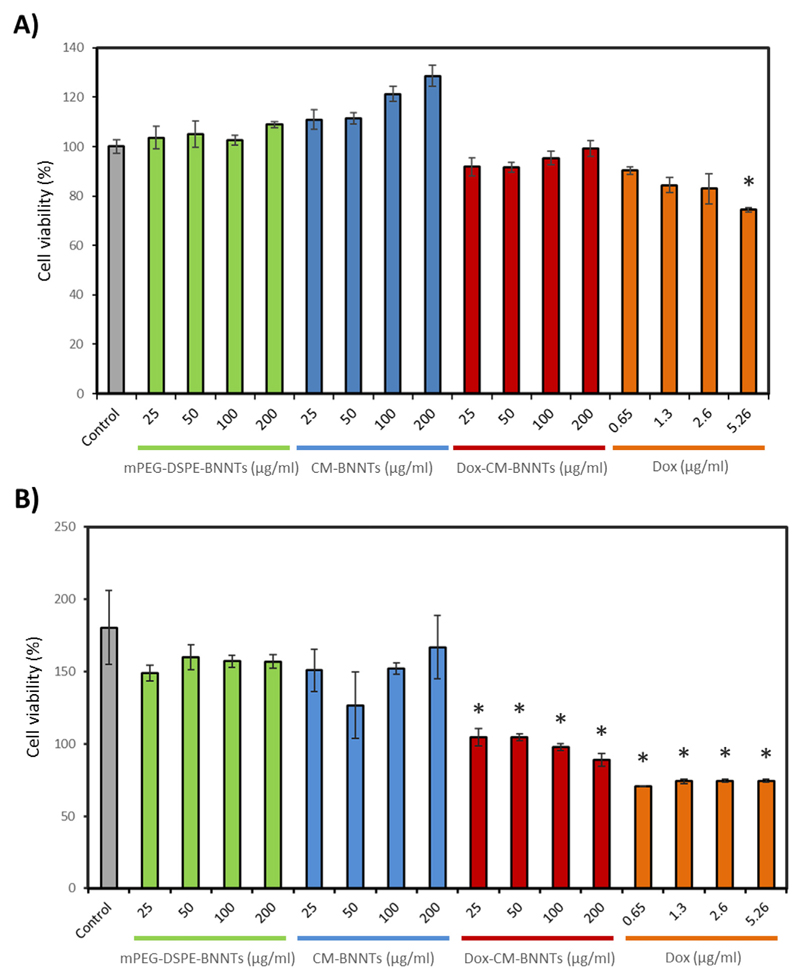
The anti-cancer properties of Dox-CM-BNNTs at different concentrations (25, 50, 100, and 200 μg/ml) were investigated on U87 MG cells and compared to cytotoxicity of CM-BNNTs and mPEG-DSPE-BNNTs (Fig. 11). Results have been moreover compared to treatments with 0.65, 1.30, 2.60, and 5.26 μg/ml of free Dox, that correspond to the drug concentrations loaded in 25, 50, 100 and 200 μg/ml of Dox-CM-BNNTs, respectively. The viability assays indicated a low, yet significant, decrement of metabolic activity for 5.26 μg/ml of free drug after 24 h (Fig. 11A). At 72 h, no significant effects were observed when exploiting plain BNNTs, both in case of CM-BNNTs and mPEG-DSPE-BNNTs, at all tested concentrations (Fig. 11B). This result demonstrates the safety and the biocompatibility of the nanocarriers, and it is in line with other works in the literature where low BNNT cytotoxicity is reported [88].
Fig. 11.
Cell viability assay on U87 MG cells treated with plain nanocarriers (mPEG-DSPE-BNNTs or CM-BNNTs), Dox-CM-BNNTs, and free doxorubicin for A) 24 h and B) 72 h (* p < .05).
Dox-CM-BNNTs and free drug were able to significantly decrease the cell viability with respect to non-treated controls and to BNNT controls (p < .05). This observation highlights the remarkable anti-cancer effects of Dox-CM-BNNTs; indeed, when cultures are treated with 100 μg/ml of Dox-CM-BNNTs, a remarkable decrease of cell viability was induced with respect to non-loaded 100 μg/ml CM-BNNTs (54.1% reduction); the anti-cancer effects of 100 μg/ml Dox-CM-BNNTs were comparable to those induced by 2.60 μg/ml of free Dox, which caused a 49% viability reduction compared to non-treated controls.
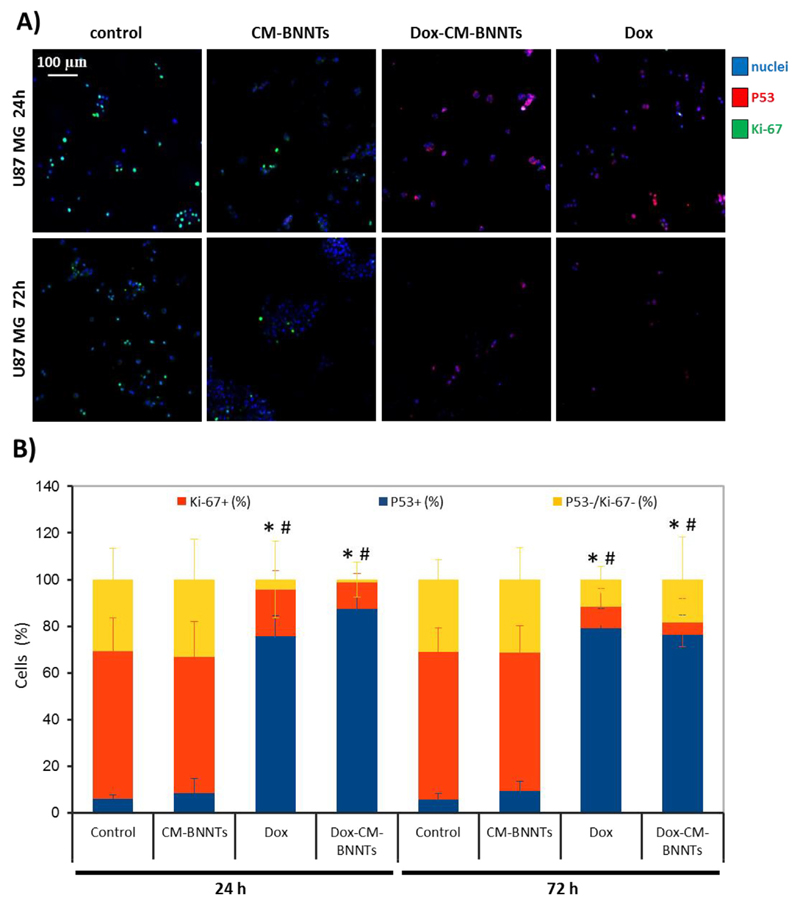
Immunocytochemistry analysis of the Ki-67 proliferation marker and of the P53 apoptotic factor were carried out on U87 MG cultures incubated with 100 μg/ml CM-BNNTs, 100 μg/ml Dox-CM-BNNTs, or 2.60 μg/ml of free Dox; results were compared to non-treated controls (Fig. 12). In Fig. 12A, the confocal fluorescence images of the different experimental classes are shown at 24 and 72 h of treatment; the percentage of Ki-67-positive nuclei and of P53-positive nuclei are reported in the graph of Fig. 12B. The negative control for immunostaining (without primary antibody) in Dox-treated samples (2.60 μg/ml) is presented in Fig. S7, and shows no significant fluorescence signal by using the scanning conditions adopted for Ki-67 and P53 imaging. Immunofluorescence analysis showed an elevated expression of the Ki-67 marker in controls and in CM-BNNTs-treated cells at both time points (at 24 h, Ki-67+ nuclei in control cultures = 63.40 ± 14.21% and Ki-67+ nuclei in CM-BNNTs-treated cultures = 58.61 ± 14.91%; at 72 h, Ki-67+ nuclei in control cultures = 63.27 ± 10.30% and Ki-67+ nuclei in CM-BNNTs-treated cultures = 59.27 ± 11.27%). Ki-67+ nuclear marker is significantly decreased in cells treated with Dox-CM-BNNTs and with free Dox (at 24 h, Ki-67+ nuclei = 11.39 ± 3.80% in Dox-CM-BNNTs-treated cultures; Ki-67+ nuclei = 19.94 ± 8.23% in Dox-treated cultures; at 72 h, Ki-67+ nuclei = 5.2 ± 10.41% in Dox-CM-BNNTs-treated cultures, Ki-67+ nuclei = 9.33 ± 7.80% in Dox-treated cultures). Concerning P53, a significantly higher nuclear expression was found in samples treated with Dox-CM-BNNTs and free Dox (at 24 h, P53+ nuclei = 87.53 ± 4.83% in Dox-CM-BNNTs-treated cultures and P53+ nuclei = 75.80 ± 8.58% in Dox-treated cultured; at 72 h, P53+ nuclei = 76.36 ± 8.33% in Dox-CM-BNNTs-treated cultures and P53+ nuclei = 79.03 ± 8.43% in Dox-treated cultured) with respect to controls (at 24 h, P53+ nuclei = 8.34 ± 6.36% in CM-BNNTs-treated cultures and P53+ nuclei = 5.84 ± 1.87% in non-treated cultured; at 72 h, P53+ nuclei = 9.24 ± 4.36% in CM-BNNTs-treated cultures and P53+ nuclei = 5.77 ± 2.42% in non-treated cultures). These results highlight an efficient pro-apoptotic and anti-proliferative effect of the Dox-CM-BNNT treatment, which is in line with the observed remarkable decrease of cell survival. The anti-cancer efficacy of Dox-CM-BNNTs can be attributed to toxicity of the Dox released from the nanocarriers, since no significant anti-proliferative and pro-apoptotic effects were elicited by the plain CM-BNNTs. In this regard, the elevated internalization of the CM-coated nanocarriers into the acidic organelles likely promotes a sustained release of Dox in the cells.
Fig. 12.
Immunocytochemistry investigations on Ki-67 and P53 expression in U87 MG cells treated for 24 and 72 h with CM-BNNTs, Dox-CM-BNNTs, and free Dox. A) Confocal fluorescence microscopy imaging of Ki-67 (in green), P53 (in red) and nuclei (in blue); B) graph showing the percentage of Ki-67+ and P53+ nuclei (* p < .05 for P53+ nuclei; # p < .05 for Ki-67+ nuclei). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
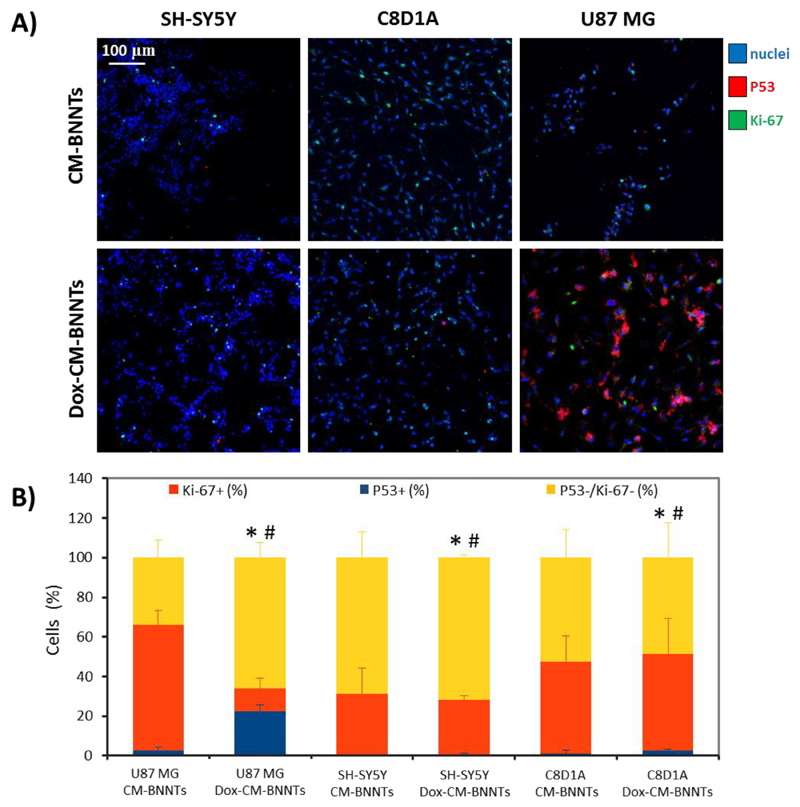
In order to evaluate the effective potential of Dox-CM-BNNTs to induce selective and targeted apoptosis, we simultaneously tested the drug-loaded nanocarriers on the three different cell lines (U87 MG, C8D1A, and differentiated SH-SY5Y), by exploiting the aforementioned home-made bioreactor. After 6 h of treatment in dynamic conditions (flow speed 1.8 ml/min), samples were washed to remove CM-BNNTs and Dox-CM-BNNTs that were non-associated to the cells, and subsequently incubated for 72 h with medium without nanotubes. Thereafter, samples were fixed and immunostained against Ki-67 and P53 markers (confocal fluorescence microscopy imaging is shown in Fig. 13A). The percentages of nuclei positive for Ki-67 (Ki-67+), P53 (P53+), or double negative for these markers (Ki-67−/P53−) are reported in the graph of Fig. 13B. A significantly higher percentage of P53+ nuclei was found in U87 MG cells treated with Dox-CM-BNNTs (22.31 ± 3.45%) with respect to the U87 MG cells incubated with CM-BNNTs (2.83 ± 1.69%; p < .05). These results, observed at 6 h of treatment in dynamic conditions, indicate as early stage apoptosis was induced [89]. Conversely, a significant decrease of Ki-67+ cells was found in U87 MG cells exposed to Dox-CM-BNNTs (11.84 ± 5.13%) compared to CM-BNNT (63.09 ± 7.21%; p < .05). Most importantly, the P53 and Ki-67 expression was not significantly affected by the Dox-CM-BNNT treatment in the other cell lines. Specifically, the percentage of P53+ cells resulted extremely low after dynamic treatment with CM-BNNTs F SH-SY5Y derived neurons, P53+ cells resulted 0.1 ± 0.2% and 0.7 ± 0.4%, respectively; for C8D1A, P53+ cells resulted 1.0 ± 1.7% and 2.60 ± 0.5%, respectively), and Ki-67 proliferation marker was not significantly affected after treatment with Dox-CM-BNNT with respect to CM-BNNTs-treated controls (for SH-SY5Y derived neurons, Ki-67+ cells resulted 31.2 ± 12.8% and 27.6 ± 1.9%, respectively; for C8D1A, Ki-67+ cells resulted 46.5 ± 12.1% and 48.6 ± 18.1%, respectively; p < .05). This result can be attributed to the scarce internalization level of the CM-BNNTs in the healthy cell lines; moreover, it demonstrates the specific anti-cancer efficacy of Dox-CM-BNNTs against GBM cells and the extremely relevant potential of the homotypic targeting approach in reducing the chemotherapy-induced side effects (54.1% reduction of cell viability in GBM cells versus no significant apoptotic effects in SH-SY5Y and C8D1A control cell lines).
Fig. 13.
Selective apoptosis of U87 MG cancer cells elicited by Dox-CM-BNNTs in dynamic conditions. A) Immunofluorescence imaging of Ki-67 (in green) and P53 (in red) markers in SH-SY5Y derived neurons, C8D1A astrocytes, and U87 MG cells treated with CM-BNNTs or with Dox-CM-BNNTs in dynamic conditions; B) graph showing the percentage of Ki-67+ and P53+ nuclei in the different experimental conditions (* p < .05 for P53+ nuclei; # p < .05 for Ki-67+ nuclei). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Similar results on homotypic targeting and selective death have been recently obtained by Wang et al. on melanoma cells [90]. In this work, a complex hybrid nanosystem has been obtained by coating Dox-loaded hollow copper sulfide nanoparticles with a mix of membranes derived from red blood cells and B16-F10 melanoma cells fused together. This nanosystem (CuS@[RBC—B16]) resulted efficiently and selectively internalized by B16-F10 cells; specifically, a 7/9-folds higher fluorescence intensity was detected in B16-F10 melanoma cells with respect to other cell lines (HT1080, NHDF, and A549 cells) upon 4 h of incubation with DiI-stained CuS@[RBC—B16]. Despite Dox-loaded CuS@[RBC—B16] displayed an anti-cancer efficacy (24.2% of cells death) lower with respect to our Dox-loaded CM-BNNTs (54.1% reduction of cell viability), authors exploited the near infra-red photothermal conversion property of CuS for obtaining a synergic hyperthermal and chemotherapy treatment of melanoma cells (resulting into a 94.5% of cell death). In the context of combined physico-chemical anti-cancer treatments, it is worth mentioning that BNNTs can also be exploited as a boron reservoir in boron neutron capture therapy (BNCT) [91]. This anti-cancer approach, that exploits epithermal neutron beam radiation, has been already successfully tested in clinical trials, with improved survival in patients with newly diagnosed glioblastoma (mean overall survival of 20.7 months with 2 years survival of 25% for patients treated with BNCT) [92]. 10B-enriched CM-BNNTs would, therefore, represent a double carrier of both B and Dox; future works may be devoted to investigate the synergic effect of BNCT and chemotherapy for the treatment of glioblastoma multiforme by taking advantage of the outstanding homotypic targeting efficacy in precision medicine.
Another remarkable example of nanoparticle targeting to cancer cells via homotypic recognition is represented by Dox-loaded gold nanocages (AuNs) coated with 4T1 CM, that have been exploited by Sun et al. for inhibiting the growth of breast cancer [93]. Similarly to the work of Wang et al., in this research a photothermal therapy was carried out by exploiting near infra-red radiation in order to induce hyperthermia and to enhance the anti-cancer efficacy. Interestingly, the combined stimulation was able not only to significantly reduce the volume of the tumor mass, yet also to decrease the metastatic capability of breast cancer in terms of number of pulmonary metastatic nodules.
Other nanotechnology-based approaches adopting cancer cell self-recognition for selective cell death also include CM-encapsulated magnetic nanoparticles loaded with Dox, CM-cloaked Dox with poly(lactic-co-glycolic acid) nanoparticles for the treatment of hepatocellular carcinoma, and paclitaxel-loaded CM-coated polymeric nanoparticles for the therapy of breast cancer and of its metastasis [27,94,95].
Obviously, the excellent targeting capabilities deriving from homotypic recognition can also be exploited for improving the diagnostic capabilities of different nanoprobes. In this specific context, the group of Pang demonstrated the bioimaging in the second near infrared window of breast cancer in mice by using Ag2Te quantum dots encapsulated within cancer CM; specifically, improved nanoprobe accumulation at tumor sites and higher tumor-to-normal tissue signal ratio were demonstrated thanks to the CM coating [96].
Finally, it is worth some consideration about the size and the non-biodegradability of the proposed nanoplatform. First of all, we have to highlight as biodistribution and pharmacokinetics of nanoparticles with sizes around 300 nm may generate concerns when intravenously administered, due to filtration and accumulation in the liver with consequent reduced circulation half-life, decreasing the chances of reaching the tumor site [97,98]. Moreover, no (or scarce) biodegradation of BNNTs can arise problems of long-term accumulation and inflammatory response. This notwithstanding, several studies by different groups, including ours, have shown satisfactory results in terms of biodistribution, circulation half-life, and biocompatibility [99,100] of BNNT-based systems owning similar sizes, all features that make these tubular structures excellent candidates for theranostic applications. Moreover, in our work we demonstrated as CM-BNNTs selectively target tumor cells, with high efficiency and with very low uptake by the other cell lines: results that strongly suggest reduced undesired effects originated by aspecific accumulation.
4. Conclusion
Specific targeting of cancer cells represents an important challenge for nanomedicine to promote tumor regression by reducing the side effects of anti-cancer drugs. In this context, homotypic recognition is a new promising targeting strategy that exploits the self-recognition properties of cancer cells. In this work, we developed a new nanoplatform based on Dox-CM-BNNTs for the targeting and treatment of glioblastoma cells in vitro. The proposed nanocarriers resulted in being an efficient drug delivery systems characterized by excellent in vitro BBB crossing, targeting efficiency, and pH-dependent release properties. The proteomic analysis allowed identifying the key role of cadherin-2 (CDw-325) and neuroplastin (SDR-1) in homotypic recognition of CM-BNNTs by glioblastoma cells. Finally, strong anti-cancer efficacy of Dox-CM-BNNTs in U87 MG cells, without significant side effects in healthy cells, was demonstrated in a complex in vitro multi-cellular and dynamic model. Altogether, these results demonstrated as the proposed nanoplatform is a promising candidate in anti-cancer therapy. This is of course a first step toward actual therapeutic applications, and we are conscious that indeed in vivo testing will be necessary before claiming success of such approach. Future works will be thus devoted to investigate the blood-brain barrier crossing efficiency of CM-BNNTs and to validate their targeting efficiency on in vivo models of glioblastoma multiforme.
Supplementary Material
Supplementary data to this article can be found online at https://doi. org/10.1016/j.matdes.2020.108742.
Highlights.
First example of a biomimetic camouflage of boron nitride nanotubes
Evidence of homotypic targeting of boron nitride nanotubes
Extensive proteomics investigations describing the homotypic targeting
Systematic investigations on in vitro complex systems, including multicellular fluidic models and blood-brain barrier models
Acknowledgments
The authors would like to thank Mr. Paolo Faraci and Dr. Pasqualantonio Pingue, Scuola Normale Superiore, Pisa (Italy) for their assistance in the ultracentrifuge facilities.
Funding sources
This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement N°709613, SLaMM).
Footnotes
CRediT authorship contribution statement
Daniele De Pasquale: Investigation, Writing - review & editing. Attilio Marino: Investigation, Writing - review & editing. Christos Tapeinos: Investigation, Writing - review & editing. Carlotta Pucci: Investigation, Writing - review & editing. Silvia Rocchiccioli: Formal analysis, Writing - review & editing. Elena Michelucci: Formal analysis, Writing - review & editing. Francesco Finamore: Formal analysis, Writing - review & editing. Liam McDonnell: Formal analysis, Writing - review & editing. Alice Scarpellini: Software, Writing - review & editing. Simone Lauciello: Software, Writing - review & editing. Mirko Prato: Formal analysis, Writing - review & editing. Aitor Larrañaga: Formal analysis, Writing - review & editing. Filippo Drago: Formal analysis, Writing - review & editing. Gianni Ciofani: Conceptualization, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability statement
The data required to reproduce these findings are available from the Corresponding Authors at resonable request.
References
- [1].Saadeh F, El Iskandarani S, Najjar M, Assi HI. Clin Neurol Neurosurg. 2019;182:98. doi: 10.1016/j.clineuro.2019.05.008. [DOI] [PubMed] [Google Scholar]
- [2].Cornillie J, Wozniak A, Vreys L, Wellens J, Pokreisz P, Hompes D, Debiec-Rychter M, Sciot R, Schoffski P. J Clin Oncol. 2015;33:e13539. [Google Scholar]
- [3].Merker PC, Lewis MR, Walker MD, Richardson EP. Toxicol Appl Pharmacol. 1978;44:191. doi: 10.1016/0041-008x(78)90298-3. [DOI] [PubMed] [Google Scholar]
- [4].Neuwelt EA, Pagel M, Glassberg M, Frenkel EP. Cancer Res. 1981;41:4466. [PubMed] [Google Scholar]
- [5].Stan AC, Casares S, Radu D, Walter GF, Brumeanu TD. Anticancer Res. 1999;19:941. [PubMed] [Google Scholar]
- [6].Lin R, Ng LS, Wang CH. Biomaterials. 2005;26:4476. doi: 10.1016/j.biomaterials.2004.11.014. [DOI] [PubMed] [Google Scholar]
- [7].Saul JM, Annapragada A, Natarajan JV, Bellamkonda RV. J Control Release. 2003;92:49. doi: 10.1016/s0168-3659(03)00295-5. [DOI] [PubMed] [Google Scholar]
- [8].Steiniger SCJ, Kreuter J, Khalansky AS, Skidan IN, Bobruskin AI, Smirnova ZS, Severin SE, Uhl R, Kock M, Geiger KD, Gelperina SE. Int J Cancer. 2004;109:759. doi: 10.1002/ijc.20048. [DOI] [PubMed] [Google Scholar]
- [9].Cerna T, Stiborova M, Adam V, Kizek R, Eckschlager T. J Cancer Metastasis Treat. 2016;2:407. [Google Scholar]
- [10].Martinelli C, Pucci C, Ciofani G. APL Bioeng. 2019;3 doi: 10.1063/1.5079943. 011502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rekha MR, Sharma CP. Peptide and Protein Delivery. 2011;165 [Google Scholar]
- [12].Teleanu DM, Chircov C, Grumezescu AM, Volceanov A, Teleanu RI. Pharmaceutics. 2018;10:269. doi: 10.3390/pharmaceutics10040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oberoi RK, Parrish KE, Sio TT, Mittapalli RK, Elmquist WF, Sarkaria JN. NeuroOncology. 2016;18:27. doi: 10.1093/neuonc/nov164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhou Y, Peng Z, Seven ES, Leblanc RM. J Control Release. 2018;270:290. doi: 10.1016/j.jconrel.2017.12.015. [DOI] [PubMed] [Google Scholar]
- [15].Liu Z, Cai W, He L, Nakayama N, Chen K, Sun X, Chen X, Dai H. Nat Nanotechnol. 2007;2:47. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- [16].Park J, Wrzesinski SH, Stern E, Look M, Criscione J, Ragheb R, Jay SM, Demento SL, Agawu A, Licona Limon P, Ferrandino AF. Nat Mater. 2012;11:895. doi: 10.1038/nmat3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Adlakha-Hutcheon G, Bally MB, Shew CR, Madden TD. Nat Biotechnol. 1999;17:775. doi: 10.1038/11710. [DOI] [PubMed] [Google Scholar]
- [18].Ciofani G, Danti S, Genchi GG, Mazzolai B, Mattoli V. Small. 2013;9:1672. doi: 10.1002/smll.201201315. [DOI] [PubMed] [Google Scholar]
- [19].Chen X, Wu P, Rousseas M, Okawa D, Gartner Z, Zettl A, Bertozzi CR. J Am Chem Soc. 2009;131:890. doi: 10.1021/ja807334b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].şen Ö, Emanet M, Çulha M. Boron Nitride Nanotubes in Nanomedicine. 2016;41 doi: 10.2217/nnm-2016-0322. [DOI] [PubMed] [Google Scholar]
- [21].Lee CH, Zhang D, Yap YK. J Phys Chem C. 2012;116:1798. [Google Scholar]
- [22].Li X, Golberg D. Boron Nitride Nanotubes in Nanomedicine. 2016;79:94. [Google Scholar]
- [23].Emanet M, Sen Ö, Çulha M. Nanomedicine. 2017;12:797. doi: 10.2217/nnm-2016-0322. [DOI] [PubMed] [Google Scholar]
- [24].Davila J, Reyes KR, Cruz MM, Inyushin M, Kucheryavykh Y, Griebenow K, Kucheryavykh L. Neuro-Oncology. 2015;17:75. [Google Scholar]
- [25].Kim JS, Shin DH, Kim JS. J Control Release. 2018;269:245. doi: 10.1016/j.jconrel.2017.11.026. [DOI] [PubMed] [Google Scholar]
- [26].Marino A, Camponovo A, Degl’Innocenti A, Bartolucci M, Tapeinos C, Martinelli C, De Pasquale D, Santoro F, Mollo V, Arai S, Suzuki M, et al. Nanoscale. 2019;11:21227. doi: 10.1039/c9nr07976a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhu JY, Zheng DW, Zhang MK, Yu WY, Qiu WX, Hu JJ, Feng J, Zhang XZ. Nano Lett. 2016;16:5895. doi: 10.1021/acs.nanolett.6b02786. [DOI] [PubMed] [Google Scholar]
- [28].Fang RH, Hu CMJ, Luk BT, Gao W, Copp JA, Tai Y, O’Connor DE, Zhang L. Nano Lett. 2014;14:2181. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Maziveyi M, Alahari SK. Oncotarget. 2017;8:48471. doi: 10.18632/oncotarget.17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhai Y, Su J, Ran W, Zhang P, Yin Q, Zhang Z, Yu H, Li Y. Theranostics. 2017;7:2575. doi: 10.7150/thno.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shi J, Kundrat L, Pishesha N, Bilate A, Theile C, Maruyama T, Dougan SK, Ploegh HL, Lodish HF. Proc Natl Acad Sci U S A. 2014;111:10131. doi: 10.1073/pnas.1409861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Doshi N, Zahr AS, Bhaskar S, Lahann J, Mitragotri S. Proc Natl Acad Sci U S A. 2009;106:21495. doi: 10.1073/pnas.0907127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jang HS. Molecules. 2017;22:2197. [Google Scholar]
- [34].Marino A, Genchi GG, Sinibaldi E, Ciofani G. ACS Appl Mater Interfaces. 2017;9:17663. doi: 10.1021/acsami.7b04323. [DOI] [PubMed] [Google Scholar]
- [35].Rocca A, Marino A, Del Turco S, Cappello V, Parlanti P, Pellegrino M, Golberg D, Mattoli V, Ciofani G. Biochim Biophys Acta - Gen Subj. 2016;1860:775. doi: 10.1016/j.bbagen.2016.01.020. [DOI] [PubMed] [Google Scholar]
- [36].Marino A, Arai S, Hou Y, Degl’Innocenti A, Cappello V, Mazzolai B, Chang YT, Mattoli V, Suzuki M, Ciofani G. ACS Nano. 2017;11:2494. doi: 10.1021/acsnano.6b08202. [DOI] [PubMed] [Google Scholar]
- [37].Grillone A, Battaglini M, Moscato S, Mattii L, De Julián Fernández C, Scarpellini A, Giorgi M, Sinibaldi E, Ciofani G. Nanomedicine. 2019;17:727. doi: 10.2217/nnm-2018-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gonzalez-Carter D, Goode AE, Kiryushko D, Masuda S, Hu S, Lopes-Rodrigues R, Dexter DT, Shaffer MSP, Porter AE. Nanoscale. 2019;11:22054. doi: 10.1039/c9nr02866h. [DOI] [PubMed] [Google Scholar]
- [39].Sznarkowska A, Olszewski R, Zawacka-Pankau J. Postepy Hig Med Dosw (Online) 2010;20:396. [PubMed] [Google Scholar]
- [40].Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. J Immunol. 1984;133:1710. [PubMed] [Google Scholar]
- [41].Muskhelishvili L, Latendresse JR, Kodell RL, Henderson EB. J Histochem Cytochem. 2003;51:1681. doi: 10.1177/002215540305101212. [DOI] [PubMed] [Google Scholar]
- [42].Lenting K, Verhaak R, ter Laan M, Wesseling P, Leenders W. Acta Neuropathol. 2017;133:263. doi: 10.1007/s00401-017-1671-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lee CH, Zhang D, Yap YK. J Phys Chem C. 2012;116:1798. [Google Scholar]
- [44].Merlo A, Mokkapati VRSS, Pandit S, Mijakovic I. Biomater Sci. 2018;6:2298. doi: 10.1039/c8bm00516h. [DOI] [PubMed] [Google Scholar]
- [45].Mukhopadhyay S, Scheicher RH, Pandey R, Karna SP. J Phys Chem Lett. 2011;2:2422. [Google Scholar]
- [46].Zhi C, Bando Y, Tang C, Golberg D. J Am Chem Soc. 2005;127:17144. doi: 10.1021/ja055989+. [DOI] [PubMed] [Google Scholar]
- [47].Waters K, Pandey R, Karna SP. ACS Omega. 2017;2:76. doi: 10.1021/acsomega.6b00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bhattacharjee S. J Control Release. 2016;235:337. doi: 10.1016/j.jconrel.2016.06.017. [DOI] [PubMed] [Google Scholar]
- [49].Danaei M, Dehghankhold M, Ataei S, Hasanzadeh Davarani F, Javanmard R, Dokhani A, Khorasani S, Mozafari MR. Pharmaceutics. 2018;10:57. doi: 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Barr TL, Seal S. J Vac Sci Technol A Vacuum, Surfaces, Film. 1995;13:1239. [Google Scholar]
- [51].Rouxhet PG, Misselyn-Bauduin AM, Animou F, Genet MJ, Adriaensen Y, Desille T, Bodson P, Deroanne C. Surf Interface Anal. 2008;40:718. [Google Scholar]
- [52].Beamson G, Briggs D. High resolution XPS of organic polymers, the Scienta ESCA 300 database John Wiley & Sons. Biomaterials. 1992;15:318. [Google Scholar]
- [53].Dufrêne YF, Van der Wal A, Norde W, Rouxhet PG. Bacteriol J. 1997;179:1023. doi: 10.1128/jb.179.4.1023-1028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hotchen CE, Maybury IJ, Nelson GW, Foord JS, Holdway P, Marken F. Phys Chem Chem Phys. 2015 doi: 10.1039/c5cp01244a. 11260. [DOI] [PubMed] [Google Scholar]
- [55].Naumkin AV, Kraut-Vass A, Gaarenstroom SW, Powell CJ. NIST X-ray photoelectron spectroscopy database, version 4.1 (National Institute of Standards and Technology, Gaithersburg, 2012), NIST X-Ray Photoelectron Spectrosc. Database Version 4.1. 2012 [Google Scholar]
- [56].Cooper GM, Hausman RE. The Cell: A Molecular Approach. 2nd edition. 2007. [Google Scholar]
- [57].Xu S, Olenyuk BZ, Okamoto CT, Hamm-Alvarez SF. Adv Drug Deliv Rev. 2013;65:121. doi: 10.1016/j.addr.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Proc Natl Acad Sci U S A. 2005;102:2760. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Johnson RP, Il Jeong Y, Choi E, Chung CW, Kang DH, Oh SO, Suh H, Kim I. Adv Funct Mater. 2012;22:1058. [Google Scholar]
- [60].Ceña V, Játiva P. Nanomedicine. 2018;13:1513. doi: 10.2217/nnm-2018-0139. [DOI] [PubMed] [Google Scholar]
- [61].Behzadi S, Serpooshan V, Tao W, Hamaly MA, Alkawareek MY, Dreaden EC, Brown D, Alkilany AM, Farokhzad OC, Mahmoudi M. Chem Soc Rev. 2017;46:4218. doi: 10.1039/c6cs00636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Saraiva C, Praça C, Ferreira R, Santos T, Ferreira L, Bernardino L. J Control Release. 2016;235:34. doi: 10.1016/j.jconrel.2016.05.044. [DOI] [PubMed] [Google Scholar]
- [63].Morad G, Carman CV, Hagedorn EJ, Perlin JR, Zon LI, Mustafaoglu N, Park TE, Ingber DE, Daisy CC, Moses MA. ACS Nano. 2019;13:13853–13865. doi: 10.1021/acsnano.9b04397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jeanes A, Gottardi CJ, Yap AS. Oncogene. 2008;27:6920. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Perez TD, Nelson WJ. Handb Exp Pharmacol. 2004;165:3–21. doi: 10.1007/978-3-540-68170-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Guo HB, Johnson H, Randolph M, Pierce M. J Biol Chem. 2009;284:34986. doi: 10.1074/jbc.M109.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Takeichi M. Nat Rev Neurosci. 2007;8:11. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- [68].Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW, Vandyke K. BMC Cancer. 2018;18:939. doi: 10.1186/s12885-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Van Roy F, Berx G. Cell Mol Life Sci. 2008;65:3756. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Owczarek S, Berezin V. J Int Biochem Cell Biol. 2012;44:1. doi: 10.1016/j.biocel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- [71].Kiang KMY, Leung GKK. Biomed Res Int. 2018;2:1. doi: 10.1155/2018/5364973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Van Itallie CM, Aponte A, Tietgens AJ, Gucek M, Fredriksson K, Anderson JM. J Biol Chem. 2013;288:13775. doi: 10.1074/jbc.M113.466193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Itoh M, Nakadate K, Matsusaka T, Hunziker W, Sugimoto H. Genes Cells. 2018;23:546. doi: 10.1111/gtc.12598. [DOI] [PubMed] [Google Scholar]
- [74].Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. J Cell Biol. 1999;147:1351. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hanna SM, Kirk P, Holt OJ, Puklavec MJ, Brown MH, Barclay AN. BMC Biochem. 2003;4:17. doi: 10.1186/1471-2091-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hansen MDH, Kwiatkowski AV. Int Rev Cell Mol Biol. 2013;303:1. doi: 10.1016/B978-0-12-407697-6.00001-5. [DOI] [PubMed] [Google Scholar]
- [77].Anastasiadis PZ, Reynolds AB. J Cell Sci. 2000;113:1319. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- [78].Goodison S, Urquidi V, Tarin D. J Clin Pathol - Mol Pathol. 1999;52:189. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Pokutta S, Weis WI. Annu Rev Cell Dev Biol. 2007;23:237. doi: 10.1146/annurev.cellbio.22.010305.104241. [DOI] [PubMed] [Google Scholar]
- [80].Gottardi CJ, Gumbiner BM. Curr Biol. 2001;11:792. doi: 10.1016/s0960-9822(01)00473-0. [DOI] [PubMed] [Google Scholar]
- [81].Whitmarsh AJ. Elife. 2013;2 doi: 10.7554/eLife.01428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Stenmark H. Nat Rev Mol Cell Biol. 2009;10:513. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- [83].Agarwal V, Naskar P, Agasti S, Khurana GK, Vishwakarma P, Lynn AM, Roche PA, Puri N. Biochim Biophys Acta - Mol Cell Res. 2019;1866:1618. doi: 10.1016/j.bbamcr.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Chen Y, Gan BQ, Tang BL. J Cell Physiol. 2010;225:326. doi: 10.1002/jcp.22286. [DOI] [PubMed] [Google Scholar]
- [85].Müller SL, Portwich M, Schmidt A, Utepbergenov DI, Huber O, Blasig IE, Krause G. J Biol Chem. 2005;280:3747. doi: 10.1074/jbc.M411365200. [DOI] [PubMed] [Google Scholar]
- [86].Zhitnyak IY, Bychkov IN, Sukhorukova IV, Kovalskii AM, Firestein KL, Golberg D, Gloushankova NA, Shtansky DV. ACS Appl Mater Interfaces. 2017;9 doi: 10.1021/acsami.7b08713. 32498. [DOI] [PubMed] [Google Scholar]
- [87].Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ciofani G, Danti S, D’Alessandro D, Moscato S, Menciassi A. Biochem Biophys Res Commun. 2010;394:405. doi: 10.1016/j.bbrc.2010.03.035. [DOI] [PubMed] [Google Scholar]
- [89].Fridman JS, Lowe SW. Oncogene. 2003;22:9030. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- [90].Wang D, Dong H, Li M, Cao Y, Yang F, Zhang K, Dai W, Wang C, Zhang X. ACS Nano. 2018;12:5241. doi: 10.1021/acsnano.7b08355. [DOI] [PubMed] [Google Scholar]
- [91].Ferreira HT, de Sousa MEB. Boron Nitride Nanotubes in Nanomedicine. 2016;95 [Google Scholar]
- [92].Miyatake SI, Kawabata S, Yokoyama K, Kuroiwa T, Michiue H, Sakurai Y, Kumada H, Suzuki M, Maruhashi A, Kirihata M, Ono K. J Neuro-Oncol. 2009;91:199. doi: 10.1007/s11060-008-9699-x. [DOI] [PubMed] [Google Scholar]
- [93].Sun H, Su J, Meng Q, Yin Q, Chen L, Gu W, Zhang Z, Yu H, Zhang P, Wang S, Li Y. Adv Funct Mater. 2017;27 1604300. [Google Scholar]
- [94].Liu X, Sun Y, Xu S, Gao X, Kong F, Xu K, Tang B. Theranostics. 2019;9:5828. doi: 10.7150/thno.34837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sun H, Su J, Meng Q, Yin Q, Chen L, Gu W, Zhang P, Zhang Z, Yu H, Wang S, Li Y. Adv Mater. 2016;28:9581. doi: 10.1002/adma.201602173. [DOI] [PubMed] [Google Scholar]
- [96].Zhang JJ, Lin Y, Zhou H, He H, Ma JJ, Luo MY, Zhang ZL, Pang DW. Adv Healthc Mater. 2019;8 doi: 10.1002/adhm.201900341. 1900341. [DOI] [PubMed] [Google Scholar]
- [97].Nagayama S, Ogawara K, Fukuoka Y, Higaki K, Kimura T. J Int Pharm. 2007;342:215. doi: 10.1016/j.ijpharm.2007.04.036. [DOI] [PubMed] [Google Scholar]
- [98].Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol Pharm. 2008;5:505. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Ferreira Soares DC, Ferreira TH, de Aguiar Ferreira C, Cardoso VN, de Sousa EM. J Int Pharm. 2012;423:489. doi: 10.1016/j.ijpharm.2011.12.002. [DOI] [PubMed] [Google Scholar]
- [100].Ciofani G, Danti S, Genchi GG, D’Alessandro D, Pellequer JL, Odorico M, Mattoli V, Giorgi M. J Int Nanomedicine. 2012;7:19. doi: 10.2147/IJN.S28144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.