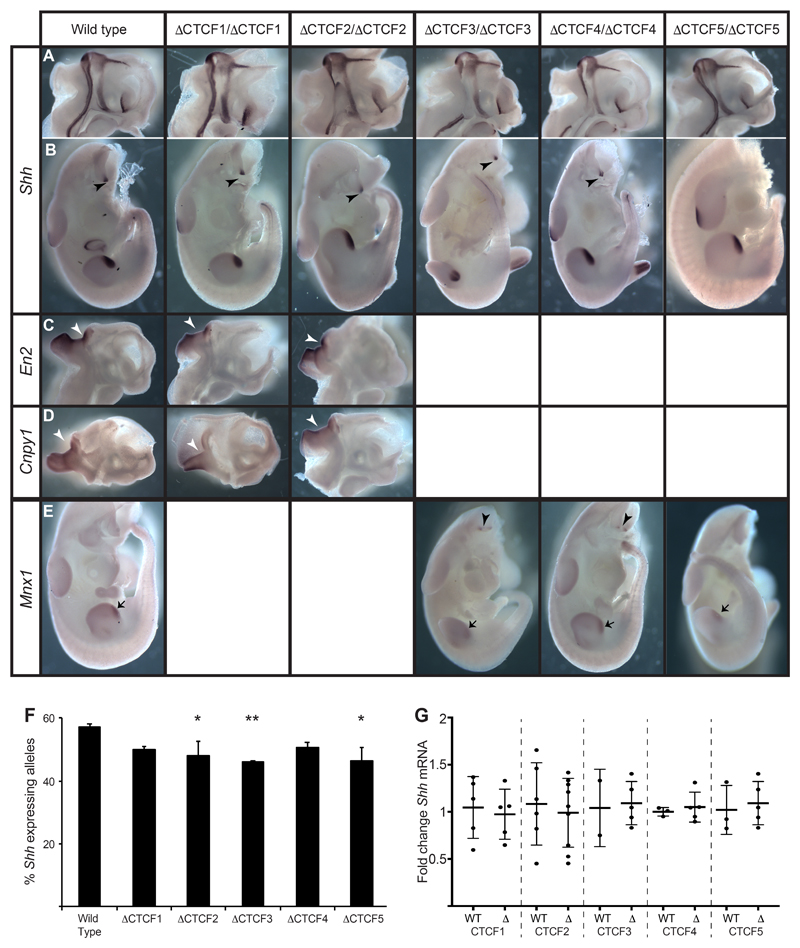
Figure 6. Expression patterns of Shh and genes in neighbouring TADs are unaffected by CTCF site deletions.
In situ analysis of gene expression at E11.5 in wild type and embryos homozygous for each of the ΔCTCF lines. (A & B) Normal expression of Shh in the midline of bisected heads (A) and in the bodies (B) where expression is detected in the ZPA of the limb bud and in the floorplate and notochord (arrowheads). (C & D) Expression of En2 (C) or Cnpy1 (D) in bisected heads. Expression is detected only at the mid brain hindbrain junction (arrowheads). (E) Expression of Mnx1 in the ZPAs of the limb buds (arrows) and in the motor neurons (arrowheads). (F) Percent of expressing Shh alleles detected by RNA FISH within the region of the brain where expression is controlled by SBE2. The statistical significance between data sets was examined by Fisher’s Exact Tests, * < 0.05, ** < 0.01. (G) Fold change of Shh expression determined by qRT-PCR of cDNA made from E11.5 heads. Mutants from each of ΔCTCF lines are compared to wildtype litter mates. Each dot represents a single embryo and data is graphed as mean +/- SD. The statistical significance between data sets was examined using unpaired Student t-tests.