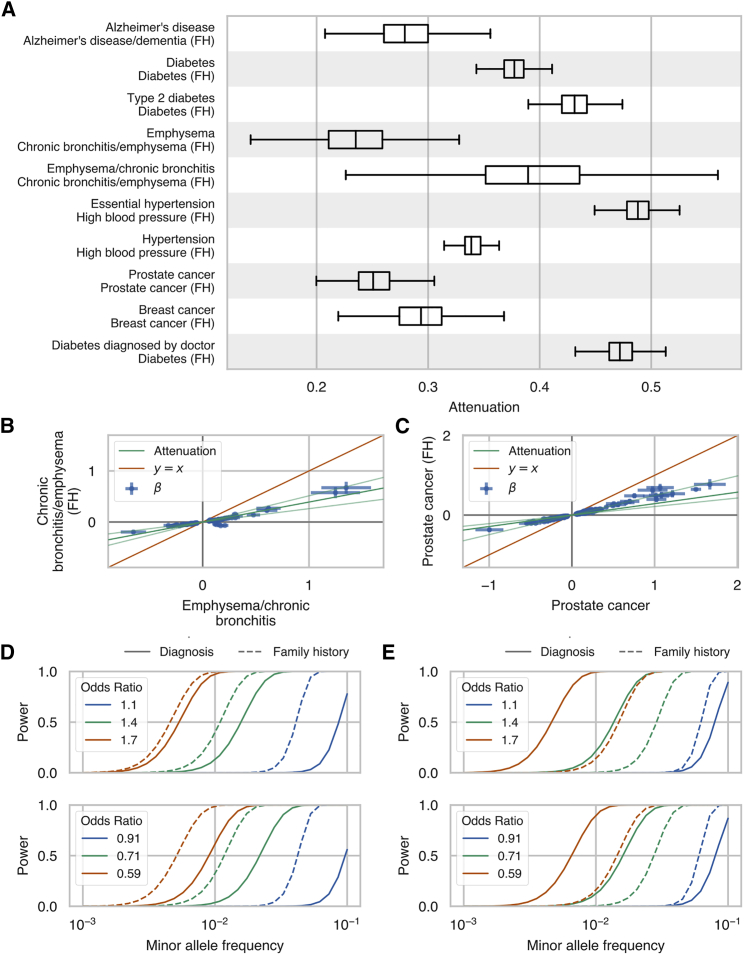
Figure 5.
GWAS Effect Size Attenuation, Scale Effects, and Power Estimates Family History GWAS
(A) Boxplots of the posterior distribution of effect size attenuation (see Material and Methods) from genome-wide association study by proxy (GWAX) using cases ascertained based on family history of disease versus genome-wide association studies (GWAS) using cases ascertained using combined hospital records and verbal questionnaire responses.
(B and C) Effect sizes and standard errors (error bars) for family history GWAX (y axis) versus combined hospital records and verbal questionnaire responses GWAS (x axis) for (B) chronic bronchitis and/or emphysema and (C) prostate cancer. The dark green line indicates the mean of the posterior distribution of attenuation from the multivariate polygenic mixture model (MVPMM) and the light green lines indicate lower and upper bounds of 95% highest posterior density of attenuation (see Material and Methods).
(D and E) Statistical power to detect association between rare genetic variants at different minor allele frequencies for (D) chronic bronchitis and/or emphysema and (E) prostate cancer in the UK Biobank. Solid lines show power for GWAS performed using cases ascertained from hospital records and questionnaire responses, and dashed lines show power for GWAS performed using cases ascertained from family history of disease. Top panel shows power for rare risk variants and bottom panel shows power for rare protective variants. Different colors indicate power for different association effect sizes. The only parameters that differ between the solid lines and dashed lines of a given color are the numbers of cases and controls.