Figure 5.
Aberrantly Processed Mutant U8 snoRNA Alleles Are Present in 29 of 33 Individuals with LCC
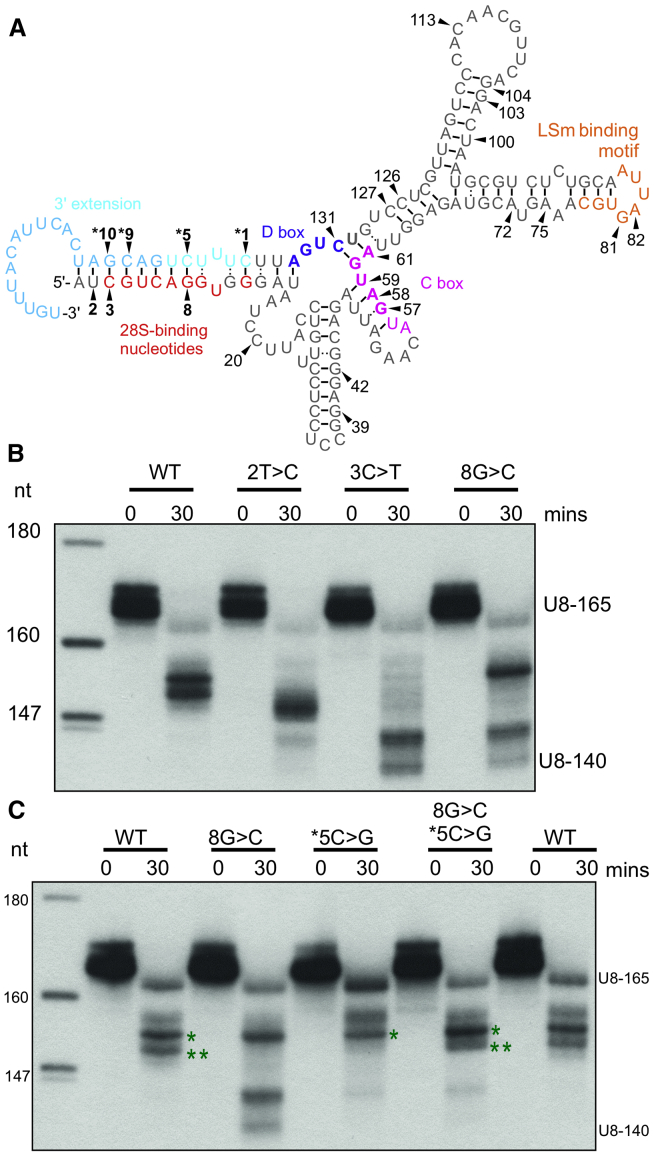
(A) Schematic depicting the secondary structure of human precursor U8 predicted by RNAfold software (18). Black arrowheads and bold numbering identify seven mutated nucleotides that lie within a duplex encompassing the 3′ extension (denoted in blue) in individuals with leukoencephalopathy with calcifications and cysts (LCC). The remaining mutated nucleotides recorded in individuals with LCC are numbered (refer to Table S1 for a complete list of LCC-associated mutant alleles). The 5′ end/3′ extension base-pairing may preclude formation of the kink-turn that is mediated by the D box (purple nucleotides) and C box (pink nucleotides) until U8 has matured sufficiently. The kink-turn interaction encompasses nucleotides 57–61 and 130–134 (bold). Important functional regions of U8 are also annotated: the nucleotides that interact with the 28S ribosomal RNA (rRNA; in red) and LSm-binding nucleotides (in orange). A black bar identifies a Watson-Crick base pairing interaction and : represents a non-Watson-Crick base pairing interaction.
(B) Processing of 5′ end-radiolabeled in vitro transcribed precursor U8 wild-type/mutant small nucleolar RNA (snoRNA; U8-165) was assessed in HeLa nuclear extracts. At 30 min, 2T>C, 3C>T, and 8G>C mutant U8 snoRNAs exhibit an enhanced rate of processing when compared to wild type as demonstrated by the presence of mature U8 (U8-140).
(C) Processing of 5′ end-radiolabeled in vitro transcribed precursor U8 wild-type/mutant snoRNA (U8-165) was assessed in HeLa nuclear extracts. At 30 min, the n∗5C>G mutant U8 is blocked in processing when compared to wild type (∗∗ band is absent), in contrast to 8G>C mutant U8 snoRNA that exhibits production of mature U8 (U8-140). When base-pairing complementarity is restored by combining the two mutations, the pattern of processing is restored to wild type.