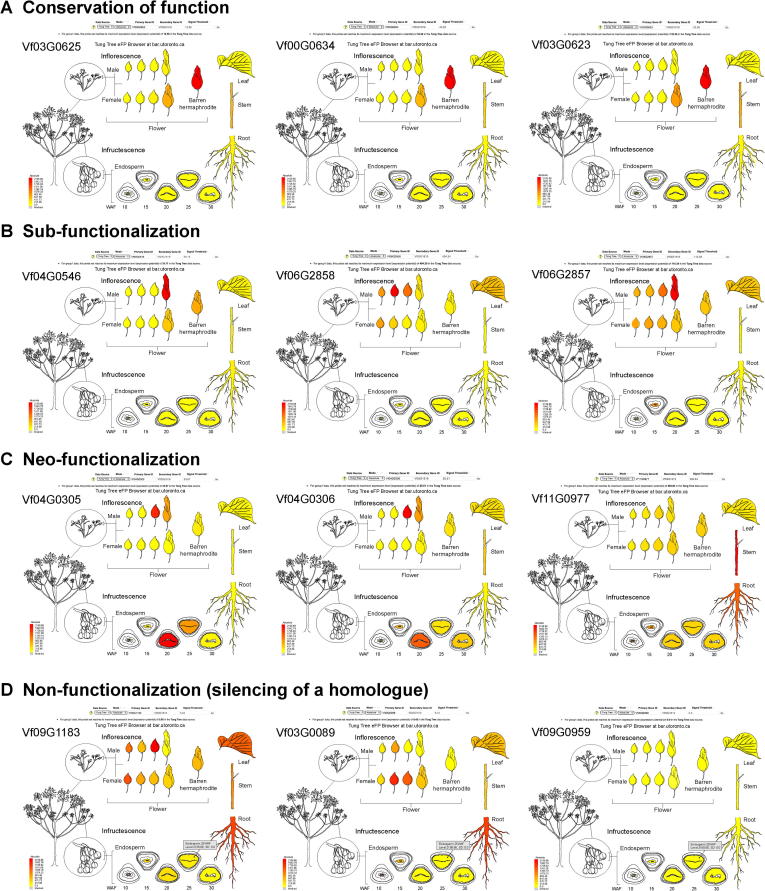
Figure 4.
Functional conservation and diversification of tung tree homologs as visualized with the tung tree eFP browser
eFP browser images showing conservation of function (A), sub-functionalization (B), neo-functionalization (C), and non-functionalization (D) of tung tree homologs. In each panel, the expression patterns of three homologs of each gene are shown. In all cases, red represents higher levels of transcript accumulation and yellow represents a lower level of transcript accumulation. From A to D, genes encoding F6′H (from left to right Vf03G0652, Vf00G0634, and Vf03G0623), protein ECERIFERUM (from left to right Vf04G0546, Vf06G2858, and Vf06G2857), PAP (from left to right Vf04G0305, Vf04G0306, and Vf11G0977), and protein LYK5 (from left to right Vf09G1183, Vf03G0089, and Vf09G0959), are shown, respectively. WAF, week after flowering; eFP, electronic fluorescent pictographic; F6′H, feruloyl CoA ortho-hydroxylase; PAP purple acid phosphatase; LYK, lysin motif receptor kinase.