Figure 1.
Genetic and gene expression divergence of primary cell cultures
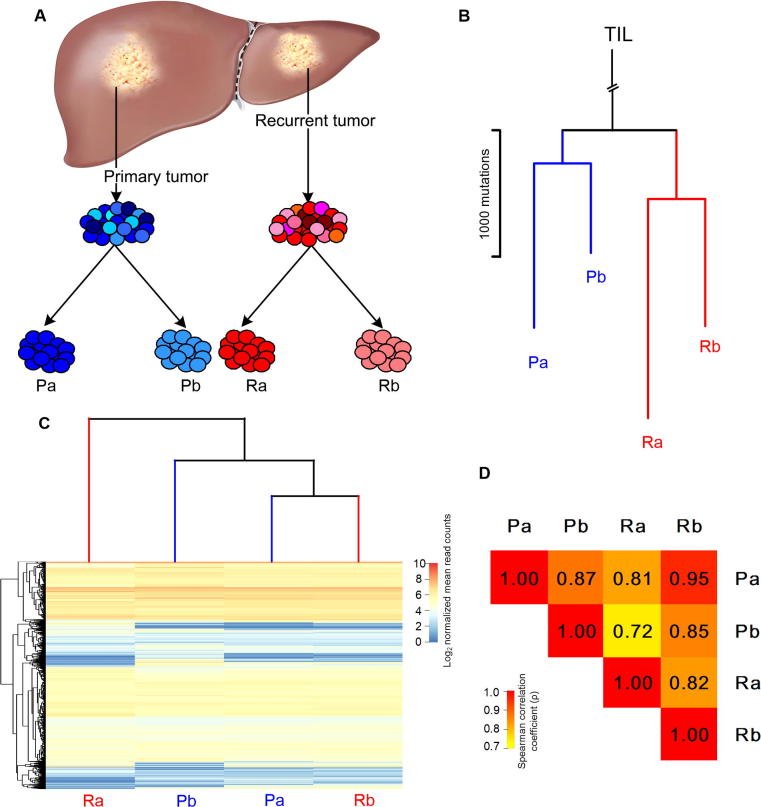
A. The four primary cell cultures from primary and recurrent tumors of an HCC patient. Primary and recurrent tumor tissues were mechanically dissociated. Primary tumor cells were initially cultured with 20% autologous serum and 10% FBS to obtain Pa and Pb, respectively. Recurrent tumor cells were initially cultured with 20% autologous serum and 10% FBS to obtain Ra and Rb, respectively. B. Phylogenetic tree based on SNVs calling from WGS data. The tree was anchored using a germline DNA sequence from TILs cultured from the recurrent tumor sample. Blue and red lines represent primary and recurrent tumors, respectively. The branch length from the (TILs) to the common ancestor is not shown to scale because of the great distance. The SNVs located in LOH regions were excluded in this analysis. C. Divergence of transcriptional profiles of cell cultures. Hierarchical clustering of the four cell cultures was performed based on normalized mean read counts (log2-transformed) for genes that were expressed (read count > 1) in at least one sample. The heatmap depicts the expression of genes on the log2 scale (rows) from each sample (columns). D. Heatmap of the Spearman correlation coefficients () of transcriptomes between any two of the four samples. Genes expressed (read count > 1) in at least one sample were used. HCC, hepatocellular carcinoma; FBS, fetal bovine serum; SNV, single nucleotide variation; WGS, whole-genome sequencing; TIL, tumor-infiltrating lymphocyte; LOH, loss of heterozygosity.