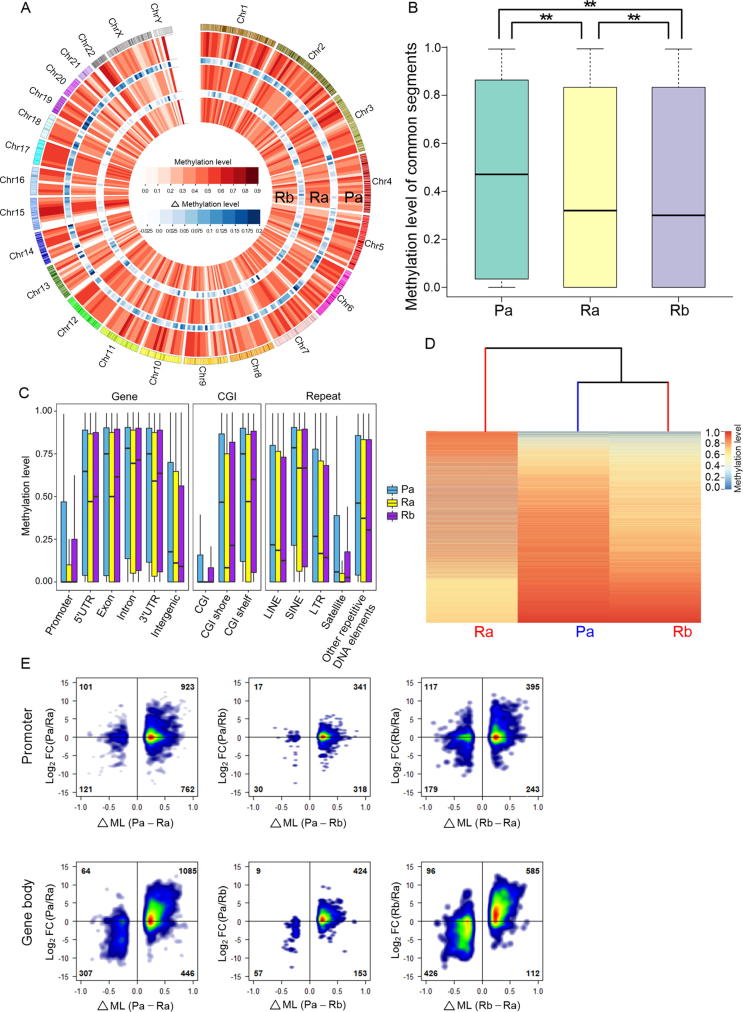
Figure 2.
Characterization of DNA methylation in Pa, Ra, and Rb
A. Circos representation of the whole-genome DNA methylation levels of the cell cultures. The outermost circle represents the chromosome locations. Different shades of red represent methylation levels averaged in 10-Mb genomic windows, and different shades of blue represent the difference in methylation levels between inner and outer layers. Inner heatmaps indicate changes in DNA methylation shown in color gradients. A gradual loss of methylation compared with that in Pa is observed (from outer to inner, Pa, Ra, and Rb). B. Boxplot of common CpG site methylation levels in cell cultures. ** indicates highly significant differences between any two samples (P < 2.2E − 16, Wilcoxon test). C. Boxplot of DNA methylation levels for multiple functional genomic categories in cell cultures. D. Divergence of the methylomes of the cell cultures. Hierarchical clustering was based on the methylation levels of differentially methylated CpG sites (≥ 10 ×) among samples. The heatmap depicts the CpG site methylation level (rows) in each sample (columns). E. Density plot of DMPs/DMGBs with the changes in the expression of associated genes between the compared samples. The horizontal and vertical axes represent differences in methylation levels (Δ ML) and expression changes (FC), respectively. Density is color-coded; red indicates a higher density and blue indicates a lower density. CGI, CpG island; LINE, long interspersed nuclear element; SINE, short interspersed nuclear element; LTR, long terminal repeat; FC, fold change; DMP, differentially methylated promoter; DMGB, differentially methylated gene body .