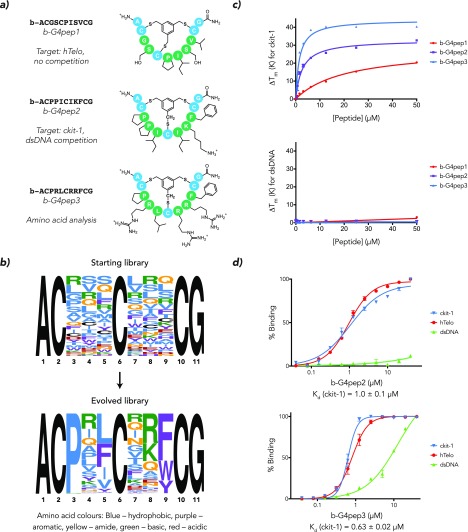
Figure 2.
(a) Schematic structures of three G4 bicyclic peptide ligands elicited by phage display, exhibiting both common G4 small molecule binding motifs and hydrophobic amino acids. (b) Next-generation sequencing permits motif analysis of the selection process; the optimal amino acids at each position in the final selected library are identified. Enriched amino acid functionalities include positive charges (position 8) and aromatic rings (9) but also hydrophobic residues (5) and proline (3). (c) ckit-1 G4 melting temperature increase (ΔTm) as measured by FRET melting. ΔTm demonstrates progressive improvement of G4 bicyclic peptides from b-G4pep1 to b-G4pep3. At the same time, no significant ΔTm is observed for a short double-stranded DNA. (d) Apparent Kd values measured by fluorescence quench equilibrium binding assay for b-G4pep1 and b-G4pep3; the quoted Kd is for ckit-1.