Figure 1.
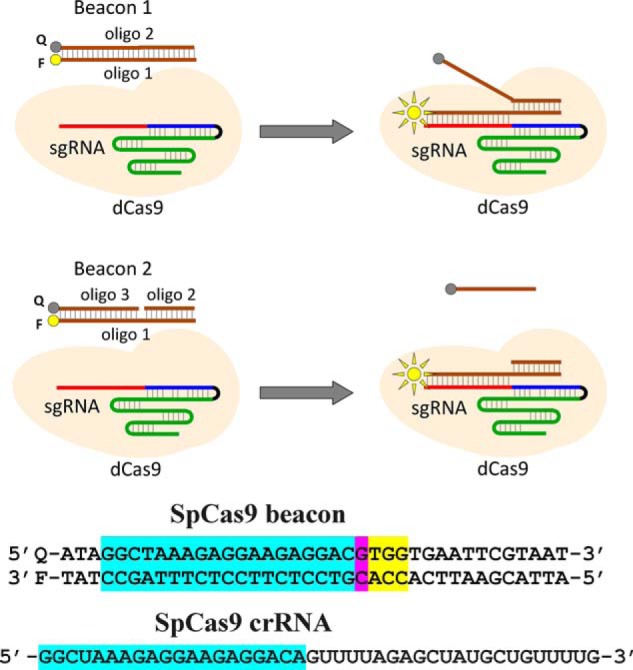
Principle of the Cas beacon assay and structure of SpCas9 beacon. Beacon 1 consists of two fully complementary oligonucleotides (oligo), whereas beacon 2 has a discontinuity in its nontarget strand. The PAM-distal ends of the beacon target and nontarget strands are labeled with a fluorophore (F) and a fluorescence quencher (Q), respectively. The baseline fluorescence intensity of beacon is low because of proximity of the fluorophore with the quencher. The Cas9–gRNA effector complex binds to beacon in a way that mimics their binding to target DNA, which leads to separation of the fluorophore and the quencher and a readily detectable increase in fluorescence intensity. The sequence of SpCas9 crRNA and beacon used in experiments with SpCas9 and are shown below the panel. The crRNA spacer and beacon protospacer bases complementary to the spacer sequence are highlighted in blue, the mismatched protospacer bases are highlighted in pink, and the PAM sequence is highlighted in yellow.
