Abstract
Optic atrophy 1 (OPA1) is a dynamin protein that mediates mitochondrial fusion at the inner membrane. OPA1 is also necessary for maintaining the cristae and thus essential for supporting cellular energetics. OPA1 exists as membrane-anchored long form (L-OPA1) and short form (S-OPA1) that lacks the transmembrane region and is generated by cleavage of L-OPA1. Mitochondrial dysfunction and cellular stresses activate the inner membrane–associated zinc metallopeptidase OMA1 that cleaves L-OPA1, causing S-OPA1 accumulation. The prevailing notion has been that L-OPA1 is the functional form, whereas S-OPA1 is an inactive cleavage product in mammals, and that stress-induced OPA1 cleavage causes mitochondrial fragmentation and sensitizes cells to death. However, S-OPA1 contains all functional domains of dynamin proteins, suggesting that it has a physiological role. Indeed, we recently demonstrated that S-OPA1 can maintain cristae and energetics through its GTPase activity, despite lacking fusion activity. Here, applying oxidant insult that induces OPA1 cleavage, we show that cells unable to generate S-OPA1 are more sensitive to this stress under obligatory respiratory conditions, leading to necrotic death. These findings indicate that L-OPA1 and S-OPA1 differ in maintaining mitochondrial function. Mechanistically, we found that cells that exclusively express L-OPA1 generate more superoxide and are more sensitive to Ca2+-induced mitochondrial permeability transition, suggesting that S-OPA1, and not L-OPA1, protects against cellular stress. Importantly, silencing of OMA1 expression increased oxidant-induced cell death, indicating that stress-induced OPA1 cleavage supports cell survival. Our findings suggest that S-OPA1 generation by OPA1 cleavage is a survival mechanism in stressed cells.
Keywords: mitochondria, mitochondrial permeability transition (MPT), necrosis (necrotic death), reactive oxygen species (ROS), oxidative stress, mitochondrial dynamics, OPA1, optic atrophy 1 (OPA1), OMA1 zinc metallopeptidase, apoptosis
Introduction
Mitochondria are essential eukaryotic organelles, supporting energetic needs of cells through oxidative phosphorylation (OXPHOS).2 Mitochondria are structurally unique, consisting of multiple compartments including the matrix and the double membranes, inner and outer membranes (IM and OM) enclosing the matrix. OXPHOS takes place in the cristae that are structurally continuous with, but functionally distinct from, the IM. Mitochondrial dynamics is a cell biological feature of mitochondria, which regulates shape, number, and location of mitochondria in cells. Fission and fusion of mitochondrial membranes are the main processes that change mitochondrial shape and size. Dynamin-family membrane-remodeling proteins mediate mitochondrial fission and fusion through GTPase activities.
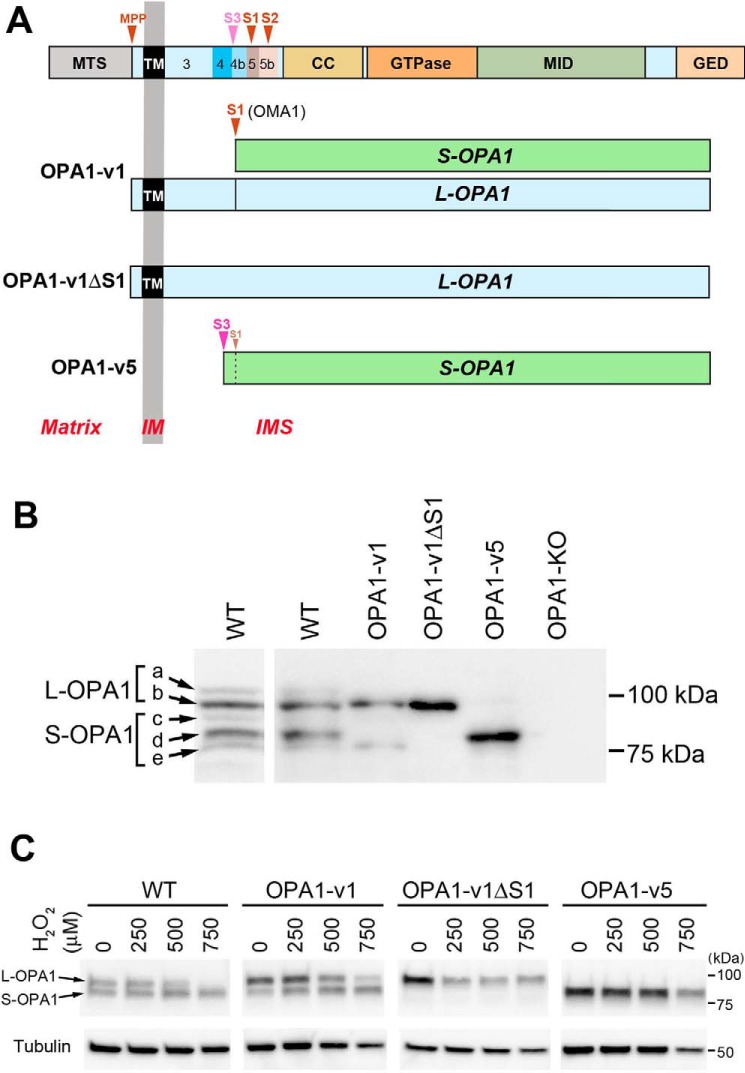
The optic atrophy 1 (OPA1) is a dynamin-related GTPase and has a profound role in mitochondrial energetics. OPA1 is associated with the IM and plays a dual role in mediating IM fusion and maintaining cristae structure, which are mechanistically independent of each other (1–6). Mutations of OPA1 decrease mitochondrial function and cause optic neuropathy as well as other systemic disorders, depending on the severity of mutation-induced mitochondrial dysfunction (7–10). Human OPA1 protein is encoded by a single gene consisting of 30 exons. Alternative splicing at the N-terminal exons 4, 4b, and 5b generates eight mRNA variants (v1–v8) (11–14). All eight precursor proteins contain the mitochondrial transit sequence at the N terminus followed by the transmembrane (TM) domain and functional enzymatic domains of dynamin proteins (GTPase domain, middle domain, and the GTPase-effector domain) (Fig. 1A). During mitochondrial import, OPA1 becomes anchored at the IM, leaving the functional domains of the protein in the intermembrane space (15). The IM-anchored OPA1 (“L-OPA1” for long form) can be cleaved downstream of the TM region, generating TM-free OPA1 (“S-OPA1” for short soluble form). It has been shown that the IM-associated metalloprotease OMA1 and the i-AAA protease Yme1L cleave OPA1 at two sites, S1 and S2, respectively, to generate five visible variants, a–e (a and b for L-OPA1; c, d, and e for S-OPA1) after electrophoretic separation (Fig. 1B, WT) (12, 13, 16–19). The S1 site is in the region encoded by exon 5 that is present in all eight OPA1 variants. In contrast, the S2 site is in the alternatively splicing exon 5b that is present in four of the eight variants. There is likely an additional cleavage site (S3 in Fig. 1A) in the region encoded by exon 4b. The S3 site is cleaved potentially by Yme1L to generate the major short form “d” (13, 17, 20, 21). The four OPA1 variants containing the protein sequence encoded by exon 4b undergo complete cleavage and thus form S-OPA1 exclusively (Fig. 1 (A and B), OPA1-v5) (13, 16, 17, 21). In the variants that do not have exon 4b, the extent of cleavage at S1 is small under normal unstressed conditions, and as a result, uncleaved L-OPA1 and a small amount of cleaved S-OPA1 are formed (OPA1-v1 in Fig. 1B). However, in stress conditions, such as mitochondrial depolarization, heat stress, and treatments with hydrogen peroxide (H2O2), oligomycin, or valinomycin, the OMA1 activity is enhanced, resulting in loss of L-OPA1 and accumulation of S-OPA1 (17–19, 22). In contrast, Yme1L-mediated OPA1 cleavage at S2 has been suggested to be associated with an increase of OXPHOS-dependent mitochondrial fusion (23).
Figure 1.
OPA1 variants used in this study. A, OPA1 protein encoded by all exons is shown at the top. MTS, mitochondrial transit sequence; CC, coiled coil domain; GTPase, GTPase domain; MID, middle domain; GED, GTPase effector domain; MPP, mitochondrial processing peptidase, S1–S3, proteolytic cleavage sites. Exons 3–5b are indicated. OPA1-v1 contains the S1 site where partial cleavage occurs, producing a small amount of S-OPA1. OPA1-v1ΔS1 has the deletion of the S1 site in OPA1-v1, producing L-OPA1 exclusively. OPA1-v5 generates S-OPA1 exclusively because of full cleavage possibly at the S3 site. B, expression of the OPA1 variants in isolated stable lines. Cell lysates of OPA1-v1, OPA1-v1ΔS1, and OPA1-v5 along with WT and OPA1-KO were immunoblotted with anti-OPA1 antibody. A separate WT immunoblot is included for clarity of different OPA1 bands (a–e). C, OPA1 cleavage in oxidant insult. Cell lysates of WT and the OPA1 variant cells treated with 250, 500, and 750 μm H2O2 for 16 h in OXPHOS media were immunoblotted for OPA1. WT and OPA1-v1 cells show the conversion of L-OPA1 to S-OPA1, most notably in 500 and 750 μm H2O2.
It was shown that L-OPA1 is fusion-competent, but S-OPA1 is not (5, 12, 13, 24, 25). Therefore, OPA1 cleavage associated with cellular stress results in mitochondrial fragmentation, which has been considered causal for mitochondrial dysfunction and cell death (26–29). It was shown that double KO cells of OMA1 and Yme1L contain only L-OPA1 due to no OPA1 cleavage and are able to maintain normal cristae structure and render apoptotic resistance (20). Conversely, prohibitin 2 (Phb2) KO induces a loss of L-OPA1 and concomitant accumulation of S-OPA1, causing mitochondrial fragmentation and cristae disruption and rendering apoptotic sensitivity. Expression of L-OPA1 in Phb2-KO cells repaired these defects, whereas S-OPA1 did not. These observations indicated that L-OPA1 alone is sufficient to support fusion, energetics, and cell survival, whereas S-OPA1 plays no active roles in fusion and cristae maintenance; hence, OPA1 cleavage is considered detrimental to cell survival by generating nonfunctional S-OPA1 (30). However, S-OPA1 generated by cleavage contains all of the functional domains of dynamin proteins and retains the GTPase activity (31), suggestive of functional significance. Indeed, our recent studies, using cells expressing L- or S-OPA1 exclusively, demonstrated directly that S-OPA1, despite its inability to mediate mitochondrial fusion, is fully competent for maintaining cristae structure and energetic activity through its GTPase activity (5), which was supported by another study (25).
Energetic competence of S-OPA1 along with the presence of all functional domains of dynamin proteins in S-OPA1 raises a possibility that OPA1 cleavage and S-OPA1 accumulation may have a physiological role, instead of being detrimental to cell survival. In the present study, by applying oxidant insult that increases OPA1 cleavage, we show that the absence of S-OPA1 markedly increases oxidant-induced cell death under OXPHOS conditions and that S-OPA1 generation is necessary for supporting cell survival. Mechanistically, we found that L- and S-OPA1 have contrasting effects on ROS production and the sensitivity to mitochondrial permeability transition (MPT). Our data indicate that L- and S-OPA1 are inherently different in maintaining mitochondrial function, in which S-OPA1 plays a protective role under cellular stress by extending cell survival.
Results
S-OPA1 confers better cell survival under oxidant insult in glucose-free OXPHOS conditions
Oxidant insult by H2O2 treatment was shown to accumulate S-OPA1 by OPA1 cleavage (20, 22). To test the roles of L- and S-OPA1 in oxidant insult, we used our mouse embryonic fibroblast (MEF) lines selectively expressing L-OPA1, S-OPA1, or both (Fig. 1, A and B) (5). OPA1-v1 has the regions encoded by exons 4 and 5 with no 4b and 5b and thus is inefficiently cleaved at S1 and forms mostly L-OPA1 with a small amount of S-OPA1 (Fig. 1B). OPA1-v1 cells increase the S-OPA1 level upon oxidant insult by enhanced cleavage at S1 (Fig. 1, A–C). OPA1-v1ΔS1 has a deletion of the S1 site in OPA1-v1, forming L-OPA1 exclusively (Fig. 1, A–C) and thus is unable to form S-OPA1 even in oxidant stress conditions. OPA1-v5 has the regions encoded by exons 4, 4b, and 5 and thus contains the S3 and S1 sites and appears to undergo complete cleavage at S3 (Fig. 1, A and B). Therefore, OPA1-v5 cells express S-OPA1 exclusively regardless of oxidant insult (Fig. 1C).
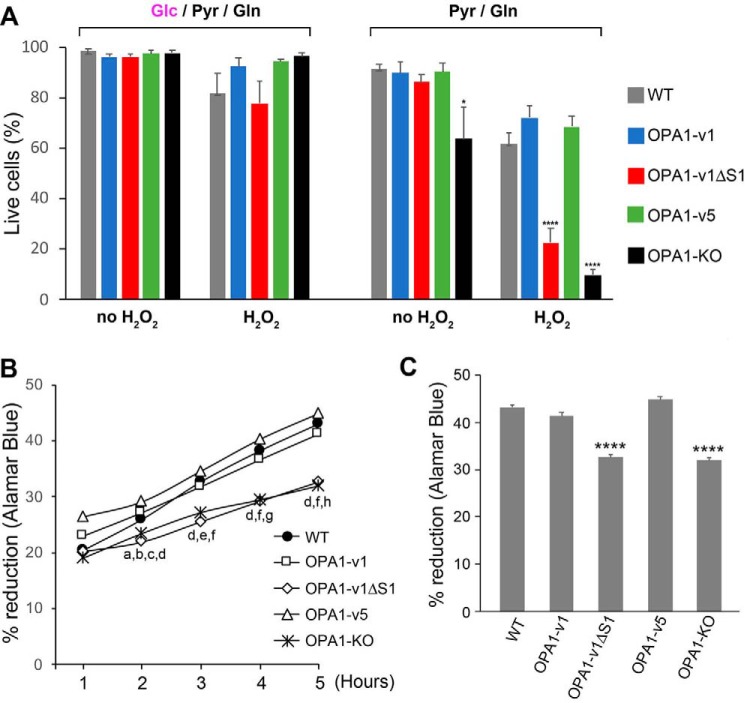
We established stable cell lines expressing each of the OPA1 variants (OPA1-v1, OPA1-v1ΔS1, or OPA1-v5) in the OPA1-KO MEFs and chose one clone for each variant, based on expression levels that are similar to the overall OPA1 level in WT cells (Fig. S1A). We treated these cells with H2O2 and examined cell survival using a trypan blue exclusion assay. We tested this in both glucose-containing and glucose-free conditions to detect potential latent difference in mitochondrial function among OPA1 variant cells. Glucose medium was the basal DMEM plus 25 mm glucose, 1 mm pyruvate, 4 mm glutamine, and 5% FBS. Glucose was omitted in glucose-free medium that essentially serves as an OXPHOS medium. After 18 h of H2O2 treatment (500 μm) in glucose medium, we found no significant death in all cell lines tested, including OPA1-KO cells (Fig. 2A), suggesting that sufficient energy by glycolysis renders cells insensitive to H2O2. Under OXPHOS conditions, without H2O2 treatment, minimal cell death was observed in WT, OPA1-v1, -v1ΔS1, and -v5 cells, indicating their similar OXPHOS capacity, consistent with our previous findings (Fig. 2A) (5). OPA1-KO cells showed an increase of cell death in OXPHOS medium, indicating defective OXPHOS activity (Fig. 2A). Unlike in glucose media, H2O2 treatment in OXPHOS conditions revealed a clear difference in cell survival between L- and S-OPA1 cells. Whereas OPA1-KO cells were most sensitive to oxidant insult as expected, the OPA1-v1ΔS1 line containing L-OPA1 exclusively was also sensitive, showing only ∼20% cell survival. Most significantly, however, S-OPA1–only cells (OPA1-v5 cells) consistently showed higher cell survival, to a similar extent as WT and OPA1-v1 cells in H2O2 treatment (60–70%) (Fig. 2A).
Figure 2.
Cells expressing L-OPA1 exclusively (OPA1-v1ΔS1) are more sensitive to oxidant insult. A, trypan blue assays of cells treated with 500 μm H2O2 for 18 h in glucose-containing and glucose-free conditions. OPA1-v1ΔS1 and OPA1-KO cells show a significant increase of cell death with H2O2 treatment in glucose-free OXPHOS conditions. n = 4 for glucose-containing; n = 6–9 for glucose-free; *, p < 0.05 versus WT and OPA1-v5 (one-way ANOVA, Turkey's multiple comparisons); ****, p < 0.0001 versus WT, OPA1-v1, and OPA1-v5 (one-way ANOVA, Turkey's multiple comparisons). Error bars, S.E. B, Alamar Blue was added after a 16-h incubation in OXPHOS media containing 500 μm H2O2, and the reduction of Alamar Blue was measured every hour for 5 h. n = 7. Comparisons within each time point are as follows. a, p = 0.001, WT versus OPA1-v1ΔS1; b, p = 0.0349, WT versus OPA1-KO; c, p = 0.0007, OPA1-v1 versus KO; d, p < 0.0001, OPA1-v1ΔS1 versus OPA1-v1 and -v5 and OPA1-v5 versus KO; e, p = 0.0004, OPA1-v1 versus KO; f, p < 0.0001, WT versus OPA1-v1ΔS1 and KO; g, p = 0.0003, OPA1-v1 versus KO; h, p < 0.0001, OPA1-v1 versus KO (two-way RM ANOVA, Turkey's multiple comparisons). C, 5-h accumulated reduction of Alamar Blue after 16-h incubation in OXPHOS media containing 500 μm H2O2. ****, p < 0.0001 versus WT, OPA1-v1, and OPA1-v5 (one-way ANOVA, Turkey's multiple comparisons).
We confirmed that the different sensitivity of the OPA1 variant cells to oxidant insult is not an artifactual effect in the particular clones selected. Incubating three different clones from each variant in glucose-free media containing 500 μm H2O2 for 24 h caused cell shrinkage and detachment in all three clones of OPA1-v1ΔS1, whereas the clones from OPA1-v1 and -v5 were well-attached and showed only a small number of dead cells (Fig. S1B), indicating that the sensitivity to oxidant insult in the OXPHOS condition is determined by differential presence of L- and S-OPA1.
We further examined cell viability by using the well-established Alamar Blue assay that measures the extent of living cell respiration by acting as an electron acceptor (32, 33). After a 16-h incubation in 500 μm H2O2 in OXPHOS media, the hourly measurements of percentage reduction of Alamar Blue showed slower rates of reduction in OPA1-v1ΔS1 and OPA1-KO cells compared with WT, OPA1-v1, and OPA1-v5 cells (Fig. 2B). The 5-h percentage reduction of Alamar Blue was significantly lower in OPA1-v1ΔS1 and OPA1-KO cells (Fig. 2C), consistent with the data from the trypan blue cell death assay. These observations that S-OPA1–containing cells (WT, OPA1-v1, and OPA1-v5 cells) survive better than L-OPA1–only cells may reflect a protective role of S-OPA1 in oxidant insult. Furthermore, the manifestation of this difference in OXPHOS conditions suggests that there may be an inherent difference between L- and S-OPA1 in maintaining mitochondrial function.
We previously reported that mitochondria in cells exclusively having either L- or S-OPA1 (OPA1-v1ΔS1 and OPA1-v5 cells, respectively) were largely fragmented, although OPA1-v1ΔS1 cells can elongate mitochondria by stress-induced mitochondrial hyperfusion (5). It has been shown that H2O2 treatment induces mitochondrial fragmentation (22). Thus, we examined mitochondrial morphology in OPA1 variant cells treated with H2O2 in OXPHOS conditions. In WT-MEFs, 500 μm H2O2 treatment increased short and fragmented mitochondria (Fig. S2), consistent with previous reports. In contrast, no significant changes were observed in OPA1-v1ΔS1 and OPA1-v5 cells treated with H2O2, which is not surprising as they intrinsically have short and fragmented mitochondria, although some increase of short tubular mitochondria (intermediate) was found with 16-h incubation in OPA1-v1ΔS1 cells (Fig. S2). Cells expressing OPA1-v1 showed a mixture of long, intermediate, and fragmented mitochondria, which was largely maintained in H2O2 incubation.
H2O2 treatment in glucose-free OXPHOS conditions induces necrotic cell death
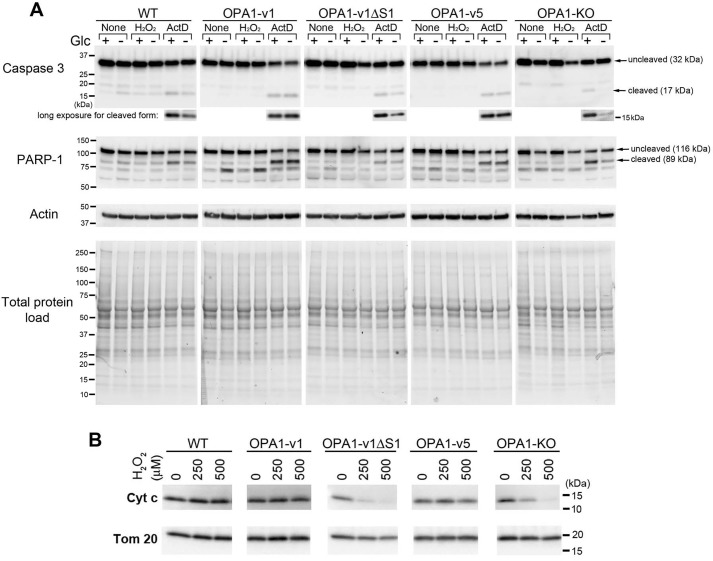
Positive trypan blue staining suggests possible necrosis by oxidant insult in OXPHOS conditions. We further tested whether H2O2-induced cell death also involves apoptosis. We examined cleavages of caspase-3 and poly(ADP-ribose) polymerase-1 (PARP-1). Activated caspase-3 by cleavage is a major executioner of apoptosis and cleaves PARP-1 (34–36). Cells were treated with H2O2 in glucose and glucose-free media. We also treated cells with a well-known apoptotic inducer, actinomycin D (actD) as a positive control for apoptosis. In all cell lines tested, actD treatment increased the cleavage products of 17 and 89 kDa for caspase-3 and PARP-1, respectively, which are known to be generated during apoptosis (37) (Fig. 3A). In contrast to actD, H2O2 treatment induced no significant generation of the 17- and 89-kDa cleavage products of caspase-3 and PARP-1 in all cell lines, including OPA1-v1ΔS1 and OPA1-KO cells that show more pronounced cell death (Fig. 2). These results suggest that H2O2-induced cell death does not involve apoptosis and is necrotic instead.
Figure 3.
H2O2 in OXPHOS conditions increases necrosis. A, immunoblotting for caspase-3 and PARP-1 of total lysates of OPA1 variant, WT, and OPA1-KO cells treated with 500 μm H2O2 or 5 μm actD with and without glucose for 16 h. Apoptotic cleavage of caspase-3 and PARP-1 is evident in actD-treated cells, but not in H2O2-treated cells. B, immunoblotting of total lysates of OPA1 variant cells for cytochrome c and Tom20 showing a loss of cytochrome c from OPA1-v1ΔS1 and OPA1-KO cells after 16-h H2O2 treatment in OXPHOS media.
Interestingly, we observed that glucose-free conditions decreased actD-induced caspase-3 cleavage compared with glucose-containing media. This decreased caspase-3 cleavage was most prominent in OPA1-KO and OPA1-v1ΔS1 cells, as well as WT cells to some extent, but was not observed in OPA1-v1 and OPA1-v5 cells (Fig. 3A, long exposure). The same trend was observed in the 89-kDa cleavage product of PARP-1. Apoptotic progression requires ATP (38). It is likely that OPA1-v1 and -v5 cells are able to produce sufficient ATP in OXPHOS conditions, allowing apoptotic progression. Despite decreased caspase-3 cleavage, we observed significant death of OPA1-KO and OPA1-v1ΔS1 cells by actD treatment in glucose-free conditions (results not shown). It is possible that OPA1-KO and OPA1-v1ΔS1 cells are unable to produce sufficient ATP in OXPHOS conditions, undergoing necrosis instead. These observations suggest that OPA1-v1 and -v5 cells may have better OXPHOS capacity than OPA1-v1ΔS1 cells, consistent with Alamar Blue data.
In addition, immunoblotting of H2O2-treated OPA1 variant cells in OXPHOS conditions revealed the loss of cytochrome c in OPA1-v1ΔS1 and OPA1-KO cells (Fig. 3B). These cells showed a significant loss of cytochrome c even in 250 μm H2O2. Because Tom20 levels were unchanged, it is likely that cytochrome c is selectively lost in these cells upon H2O2 treatment in OXPHOS media. The degradation or loss of cytochrome c from dying cells has been reported in late apoptosis and necrosis with more pronounced loss in necrotic cells (39–41). These data further support the higher necrotic sensitivity of OPA1-v1ΔS1 and OPA1-KO cells to H2O2 in OXPHOS conditions.
OPA1-v1ΔS1 cells generate more ROS than OPA1-v5 cells in glucose-free OXPHOS conditions
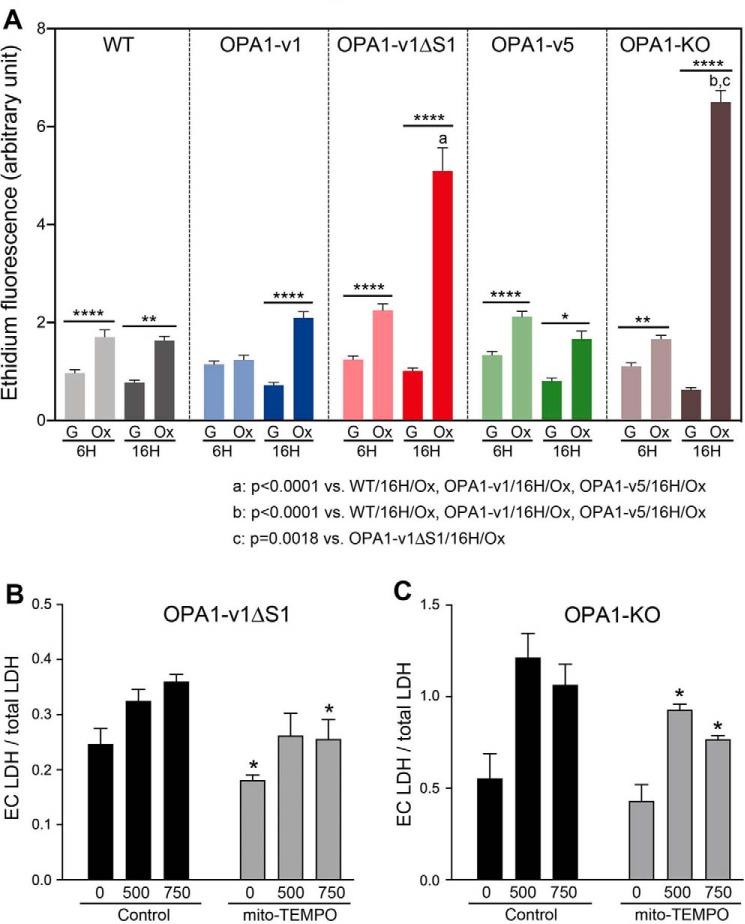
We found that the sensitivity of OPA1-v1ΔS1 cells to H2O2 is revealed only under OXPHOS conditions. Because this H2O2 sensitivity of OPA1-v1ΔS1 cells is similar to that of OPA1-KO cells (Fig. 2) that have an OXPHOS defect, there may be a latent decrease of mitochondrial function in OPA1-v1ΔS1 cells, which was not detected in previous studies (5, 25). It is possible that, under OXPHOS conditions, decreased mitochondrial function in OPA1-v1ΔS1 cells generates and accumulates more ROS over time, rendering increased sensitivity to oxidant insult. Therefore, we evaluated the ROS levels in OPA1 variant cells in OXPHOS and glucose media without H2O2 treatment. ROS levels measured by using the oxidation of dihydroethidium (DHE) indicate that, at 6 h of incubation, ROS levels were low in all cell lines in the presence of glucose (Fig. 4A). OXPHOS conditions increased ROS levels by up to 2-fold after 6-h incubation compared with glucose media. After 16 h in glucose media, ROS levels remained low. However, in OXPHOS conditions, drastic increases of ROS were observed in OPA1-v1ΔS1 and OPA1-KO cells with smaller increases in the other cell lines (Fig. 4A). Similar observations were made with MitoSOX, showing ROS increase in OPA1-v1ΔS1 and OPA1-KO cells in OXPHOS conditions (Fig. S3). The revelation of ROS increase in OPA1-v1ΔS1 cells under OXPHOS conditions may explain susceptibility of these cells to H2O2-induced cell death. These observations further suggest that there is a functional difference between L- and S-OPA1 in which S-OPA1 may play a protective role.
Figure 4.
Elevated ROS production in OPA1-v1ΔS1 and OPA1-KO cells in OXPHOS conditions. A, OPA1 variant cells along with WT and OPA1-KO cells were incubated in glucose-containing (G) or glucose-free OXPHOS (Ox) media, and superoxide levels were evaluated at 6 and 16 h by DHE. n = 106–136 for 6 h; n = 108–160 for 16 h. *, p < 0.05; **, p < 0.01; ****, p < 0.0001 (one-way ANOVA, Turkey's multiple comparisons). Error bars, S.E. B and C, mito-TEMPO decreases H2O2-induced cell death. OPA1-v1ΔS1 (B) and OPA1-KO (C) cells were treated with 500 and 750 μm H2O2 in the presence and absence of mito-TEMPO (10 μm). Released extracellular (Ec) LDH levels were evaluated for necrotic death. n = 5–6. *, p < 0.05 (Student's t test with corresponding H2O2 concentrations in control, unpaired). Error bars, S.E.
Dysfunctional mitochondria increase superoxide production. We therefore tested whether mito-TEMPO, a mitochondrial superoxide scavenger, decreases H2O2-induced necrotic cell death in OPA1-v1ΔS1 and OPA1-KO cells. For necrosis assays, we analyzed extracellular lactate dehydrogenase (LDH) levels in cells incubated with 500 and 750 μm H2O2. We found that including mito-TEMPO in H2O2 incubation decreases cell death in both OPA1-v1ΔS1 and OPA1-KO cells (Fig. 4, B and C). However, the extent of the decrease was modest, suggesting that increased superoxide production may not be the sole contributor to the increased susceptibility of these cells to oxidant-induced death.
OPA1-v1ΔS1 cells show an increase of oxidative stress in OXPHOS conditions
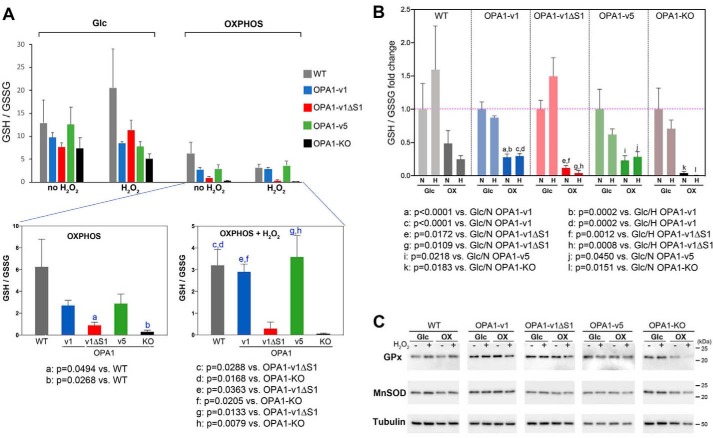
Increased ROS levels in OPA1-v1ΔS1 cells in OXPHOS conditions suggest that these cells are in oxidative stress. A decrease of cellular GSH/GSSG ratio is a measure for increased oxidative stress. We tested the GSH/GSSG ratio in OPA1 variant cells in both glucose and OXPHOS media in the presence and absence of 500 μm H2O2. Our data showed that OXPHOS conditions decreased the GSH/GSSG ratio compared with glucose conditions in all OPA1 variant cells as well as WT and OPA1-KO cells (Fig. 5, A and B). In OXPHOS conditions, OPA1-v1ΔS1 and OPA1-KO cells showed the greatest decrease in the GSH/GSSG ratio (Fig. 5, A and B). The data indicate that the presence of H2O2 in OXPHOS conditions renders more pronounced differences in GSH/GSSG ratios among OPA1 variant cells (Fig. 5A). These data indicate that OPA1-v1ΔS1 cells are under increased oxidative stress in OXPHOS conditions, compared with OPA1-v1 and -v5 cells. Immunoblotting showed that there is no significant difference in the levels of antioxidant enzymes GSH peroxidase (GPx) and Mn-superoxide dismutase (MnSOD) among OPA1 variant cells in both glucose and OXPHOS conditions regardless of the presence of H2O2 (Fig. 5C). OPA1-KO cells showed a decrease in GPx level with H2O2 in OXPHOS conditions, presumably due to cell death.
Figure 5.
Increased oxidative stress in OPA1-v1ΔS1 cells in OXPHOS conditions. Total GSH and GSSG were measured in cells treated with 500 μm H2O2 in glucose and OXPHOS media for 16 h. A, OXPHOS conditions decrease GSH/GSSG ratio compared with glucose conditions in all OPA1 variant cells, WT, and OPA1-KO cells. OPA1-v1ΔS1 and OPA1-KO cells show a greater decrease of GSH/GSSG ratio in OXPHOS conditions, which is more prominent in H2O2 treatment (bottom panels). B, fold changes of GSH/GSSG ratios showing marked decreases of GSH/GSSG ratios in OXPHOS conditions in all cells tested. Values are normalized to those of glucose with no H2O2 in respective cell lines. N, no H2O2; H, 500 μm H2O2. The data were analyzed by one-way ANOVA with Turkey's multiple comparisons. Error bars, S.E. C, immunoblotting for GPx and MnSOD shows no significant difference in all test conditions tested except for GPx in OPA1-KO cells in OXPHOS conditions.
OPA1-v1ΔS1 cells have lower respiration capacity and ATP levels than OPA1-v5 cells in oxidant insult under OXPHOS conditions
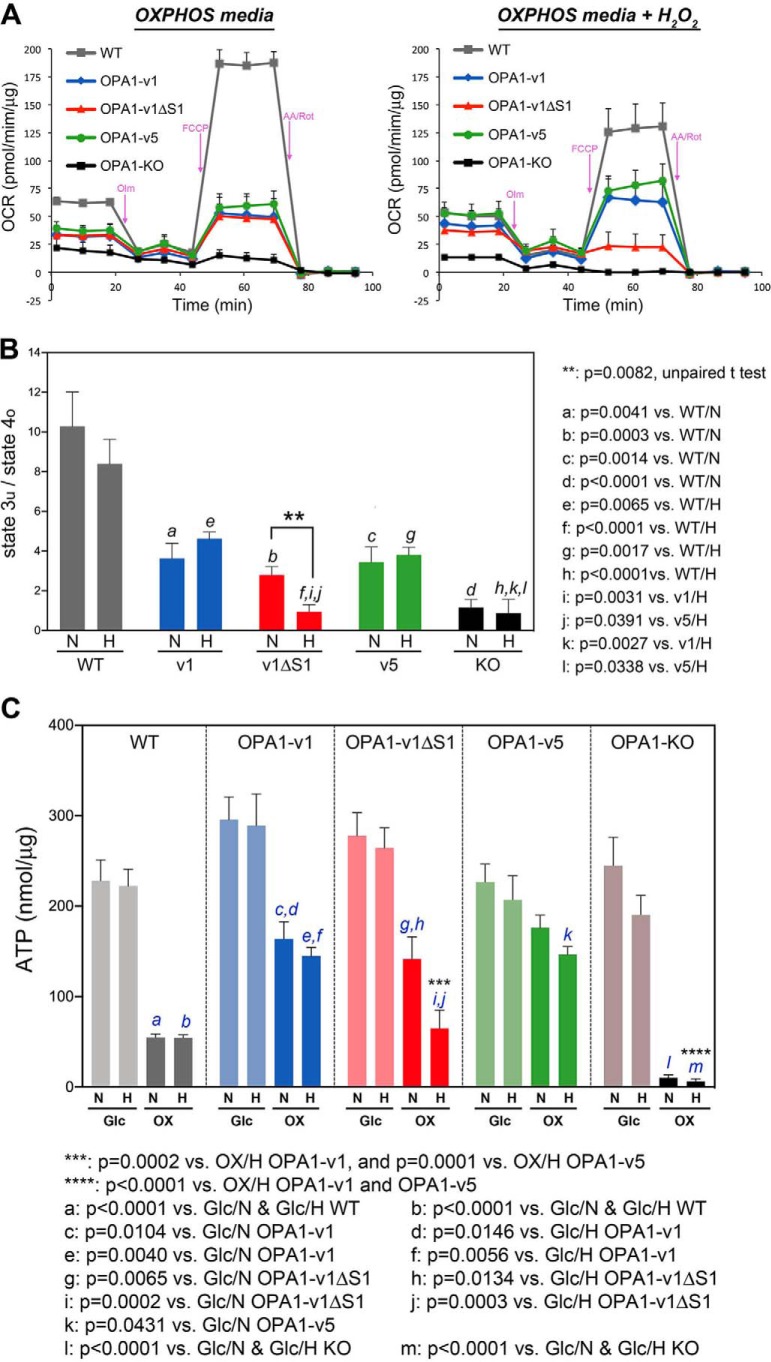
Our data demonstrated that H2O2 treatment in OXPHOS conditions increased cell death, ROS, and oxidative stress in OPA1-v1ΔS1 cells more prominently, compared with WT, OPA1-v1, and OPA1-v5 cells, suggesting that respiration of OPA1-v1ΔS1 cells may be compromised in these conditions. Indeed, respiration analyses support this notion (Fig. 6, A and B). We performed respiration analyses after incubating cells with H2O2 in OXPHOS media for 14 h, in which cells remained well-spread and attached during the assays. Our data showed that H2O2 treatment under OXPHOS conditions markedly decreased maximal respiration in OPA1-v1ΔS1 cells, but not in OPA1-v1, OPA1-v5, and WT cells, indicating that OPA1-v1ΔS1 cells are deficient in electron transport activity under these conditions (Fig. 6A). OPA1-KO cells were respiration-defective as we showed previously (5). The ratio of state 3u/state 4o, a cellular respiration control index, clearly indicates greatly decreased respiration coupling in OPA1-v1ΔS1 cells after oxidant insult in OXPHSO conditions (Fig. 6B). Immunoblotting showed no significant change in the levels of respiratory complex proteins in OPA1-v1ΔS1 cells regardless of the presence of glucose and H2O2 (Fig. S4).
Figure 6.
Oxidant insult in OXPHOS conditions decreases respiration capacity of OPA1-v1ΔS1 cells. WT, OPA1-KO, and OPA1 variant cells were treated with 500 μm H2O2 in OXPHOS media for 14 h before respiration analyses. A, OCRs of these cells. Markedly reduced maximal respiration is evident in OPA1-v1ΔS1 cells treated with H2O2. OPA1-KO cells are respiration-deficient. Error bars, S.E. B, cellular respiratory control ratios show no significant difference among the three OPA1 variant cells without H2O2 treatment. H2O2 treatment significantly decreases respiration coupling in OPA1-v1ΔS1 cells, not in OPA1-v1 and OPA1-v5 cells. N, no H2O2; H, 500 μm H2O2. One-way ANOVA was performed with Turkey's multiple comparisons. Error bars, S.E. C, ATP assays in glucose and OXPHOS conditions in the presence and absence of oxidant insult. OPA1-v1ΔS1 cells show a significantly lower ATP level than OPA1-v1 and -v5 cells in H2O2 treatment under OXPHOS conditions. N, no H2O2; H, 500 μm H2O2. One-way ANOVA was performed with Turkey's multiple comparisons. Error bars, S.E.
Our ATP assays support the respiration data, showing that OPA1-v1ΔS1 cells have a significantly lower ATP level than OPA1-v1 and -v5 cells in H2O2 treatment under OXPHOS conditions (Fig. 6C). OPA1-KO cells had very little ATP in OXPHOS conditions, as expected. Interestingly, the ATP level of WT cells treated with H2O2 in OXPHOS condition is similar to that of OPA1-v1ΔS1 cells, suggesting the involvement of other factors in cell death under oxidant insult.
Cell death induced by H2O2 in glucose-free OXPHOS conditions involves MPTs
H2O2 can induce MPT by opening of the permeability transition pore (PTP) (42, 43), causing mitochondrial dysfunction. Upon MPT, cellular energy crisis by ATP depletion causes necrotic cell death, which is the probable event for increased death of OPA1-v1ΔS1 and OPA1-KO cells by H2O2 under glucose-free conditions. Therefore, we tested the involvement of MPT in H2O2-induced cell death. Although the identity of PTP is currently unresolved, cyclophilin D (CypD) is an established regulator of PTP, and cyclosporine A (CsA) decreases MPT by inhibiting CypD (44–46). We tested whether CsA decreases necrotic death caused by H2O2 in OXPHOS conditions by evaluating extracellular LDH levels in cells incubated with 250, 500, and 750 μm H2O2.
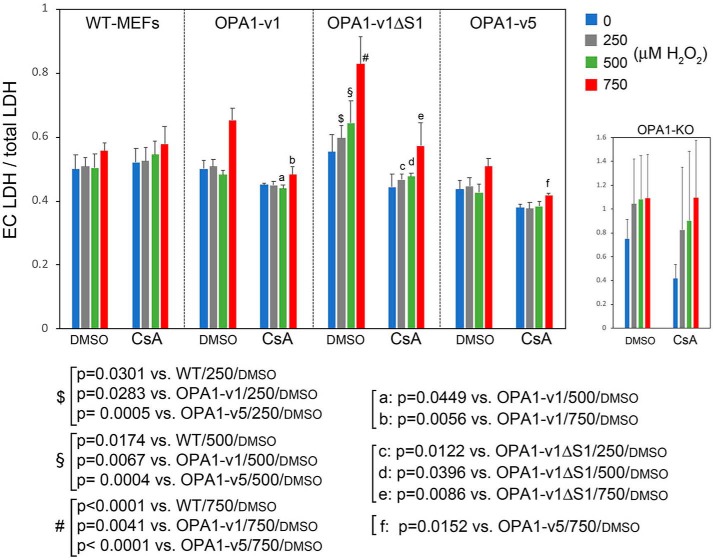
Consistent with trypan blue assays, OPA1-v1ΔS1 cells were more sensitive to H2O2 in all three H2O2 concentrations, showing significantly increased LDH release compared with WT, OPA1-v1, and OPA1-v5 cells (Fig. 7, $, §, and #). The highest H2O2 concentration (750 μm) increased LDH release in OPA1-v1 and OPA1-v5 cells as well, whereas 250 and 500 μm H2O2 did not. Importantly, in the presence of CsA, the levels of LDH release in OPA1-v1ΔS1 cells were significantly attenuated in all three H2O2 concentrations. CsA also decreased LDH release in OPA1-v1 and OPA1-v5 cells treated with 750 μm H2O2. These data indicate that MPT contributes to H2O2-induced necrosis, in which the MPT may play a larger role in the death of OPA1-v1ΔS1 cells than in OPA1-v5 cell death.
Figure 7.
H2O2-induced cell death in OXPHOS conditions involves MPT. WT and OPA1 variant cells were treated with 250, 500, and 750 μm H2O2 in OXPHOS conditions in the presence and absence of 1 μm CsA for 18 h, and cell death was evaluated by LDH assays. CsA treatment decreased cell death most prominently in OPA1-v1ΔS1 cells. n = 4–9. One-way ANOVA was performed. Error bars, S.E. Note that LDH release of OPA1-KO cells was markedly higher than other cells, although assay values vary greatly.
OPA1-v1ΔS1 cells are more sensitive to Ca2+-induced MPT than OPA1-v5 cells
We found a greater contribution of MPT to H2O2-induced necrosis in OPA1-v1ΔS1 cells than in OPA1-v5 cells (Fig. 7), suggesting that L- and S-OPA1 cells may have different sensitivities to MPT. MPT occurs through the mitochondrial Ca2+ accumulation. Therefore, we examined the MPT sensitivity of OPA1 variant cells by evaluating mitochondrial calcium retention capacity (mCRC). The mCRC was measured in digitonin-permeabilized cells by repeated additions of Ca2+ until mitochondria were unable to take up Ca2+ (47, 48). Most notably, we found that OPA1-v5 cells have the highest mCRC value (Fig. 8, A and B). On the other hand, OPA1-v1ΔS1 cells had mCRC lower than OPA1-v1 and OPA1-v5 cells, whereas OPA1-KO cells had the lowest mCRC (Fig. 8, A and B). The high and low MPT sensitivities of OPA1-v1ΔS1 and OPA1-v5 cells, respectively, agree well with the difference in cell survival of L- and S-OPA1 cells under oxidant insult. These data indicate that the higher MPT sensitivity of OPA1-v1ΔS1 and OPA1-KO cells contributes to increased cell death by oxidant insult in OXPHOS conditions.
Figure 8.
L- and S-OPA1 differentially affect the sensitivity to Ca2+-induced MPT. A, mCRC assays by repeated additions of Ca2+ to permeabilized OPA1 variant cells. Ca2+ (20 n moles each) was added to permeabilized cells (2.8 million/0.5 ml) every 2 min until no more Ca2+ was taken up. B, calculated mCRC. OPA1-v5 cells have significantly higher mCRC than OPA1-v1ΔS1 cells. n = 4–11. One-way ANOVA was performed with Turkey's multiple comparisons. Error bars, S.E. C, ROS is not involved in conferring low mCRC in OPA1-v1ΔS1 and KO cells. mCRC assays were performed with 2.9 million (OPA1- v1ΔS1) and 2.7 million cells (OPA1-KO cells) with and without antioxidants: MnTMPyP (50 μm) or mito-TEMPO (20 μm). Antioxidants had no effect on mCRC of these cells. D, the addition of H2O2 does not affect mCRC. Shown is the mCRC assay with OPA1-v1 cells (3.0 million cells) in the presence and absence of 500 μm H2O2.
ROS can potentiate MPT (42, 43), and our data show that OPA1-v1ΔS1 and OPA1-KO cells generate more ROS in OXPHOS conditions than the other cell lines (Fig. 4). Because, in the mCRC assay, mitochondria are energized with respiratory substrates, it is possible that increased ROS production from OPA1-v1ΔS1 and OPA1-KO mitochondria sensitizes these cells to Ca2+-induced MPT. To test the involvement of ROS in the increased MPT sensitivity of OPA1-v1ΔS1 and OPA1-KO cells, we included antioxidants in mCRC assays. We found that neither mito-TEMPO nor MnTMPyP improved mCRC of OPA1-v1ΔS1 and OPA1-KO cells (Fig. 8C). Furthermore, directly adding H2O2 to the mCRC assay did not decrease the MPT sensitivity in OPA1-v1 cells (Fig. 8D). Because the mCRC assay is mainly Ca2+-driven in permeabilized cells, and thus possibly bypasses ROS generation and its effect, these data indicate that ROS is not the factor that makes these cells sensitive to MPT. As such, the different MPT sensitivities of OPA1-v1ΔS1 and OPA1-v5 cells are likely the intrinsic properties exerted by L- and S-OPA1.
To further test the differential MPT in intact cells, we used the cobalt-quenching calcein assay. In this assay, intact mitochondria retain calcein fluorescence, which is quenched by cobalt ions upon MPT (49, 50). We examined mitochondrial calcein fluorescence in OPA1 variant cells treated with 500 μm H2O2 in OXPHOS conditions. We found that OPA1-v5 cells retained higher mitochondrial calcein fluorescence than other cells (Fig. S5A), consistent with OPA1-v5 rendering reduced sensitivity to MPT.
Immunoblotting showed a drastically reduced level of CypD in OPA1-KO cells compared with WT cells (Fig. S5B). The OPA1 variant cells also contained lower CypD levels than WT cells. Interestingly, the CypD levels of OPA1-v5 cells were even lower than those of OPA1-KO, OPA1-v1, and OPA1-v1ΔS1 cells (Fig. S4B), which may support the increased mCRC of OPA1-v5 cells. However, OPA1-KO cells have much lower mCRC than WT cells despite its reduced CypD level. It is likely that the CypD level is not the sole component determining the extent of mCRC. Further studies are necessary to determine the mechanism of regulating MPT in OPA1 variant cells.
Stress-induced OPA1 cleavage is beneficial for cell survival under oxidant insult
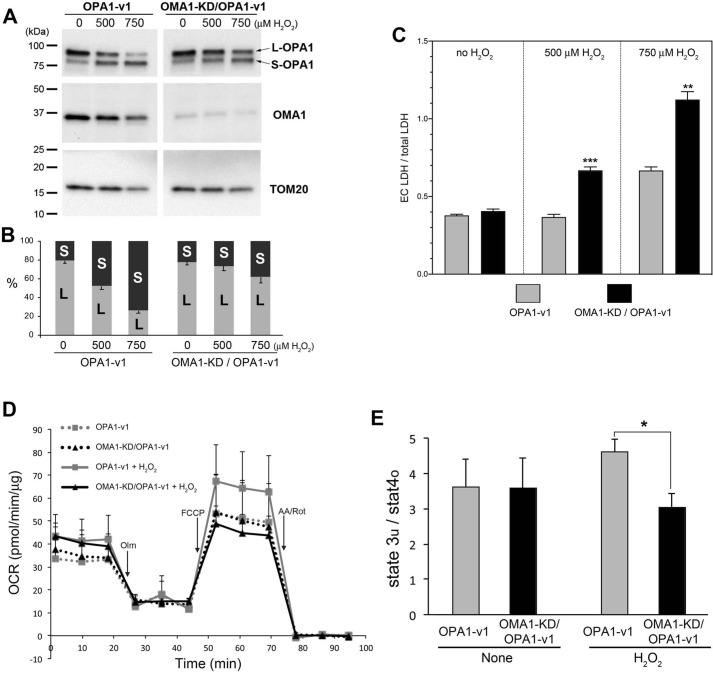
Our data show that the absence of S-OPA1 (OPA1-v1ΔS1 cells) significantly increases cell death under oxidant insult, suggesting that S-OPA1 generation may be beneficial for cell survival under stress conditions. Therefore, we hypothesized that S-OPA1 generation by OPA1 cleavage in stress conditions plays a protective role. In stress conditions, OMA1 is activated and cleaves OPA1 at the S1 site (17–19, 22). OPA1-v1 has the single cleavage site of S1, and H2O2 treatment increases the OPA1 cleavage to convert L-OPA1 to S-OPA1 (Fig. 1). To test our hypothesis, we silenced OMA1 in OPA1-v1 cells and asked whether preventing OPA1 cleavage increases cell death in oxidant insult. We generated stable cells expressing OMA1-shRNA in OPA1-v1 cells. Immunoblotting shows that OMA1 silencing in OPA1-v1 cells significantly attenuated OPA1 cleavage in H2O2 treatment (Fig. 9, A and B). LDH assays indicate that OPA1-v1 cells with silenced OMA1 show increased cell death in H2O2 treatment in OXPHOS conditions (Fig. 9C). Respiration analyses show that OMA1 silencing in OPA1-v1 cells decreased respiration after H2O2 treatment in OXPHOS conditions, indicating decreased mitochondrial function (Fig. 9, D and E). These data demonstrate that S-OPA1 generation by OPA1 cleavage is necessary for maintaining mitochondrial function and supporting cell survival in oxidant insult, providing evidence for the functional importance of S-OPA1 and its generation. Contrary to conventional notions, our data indicate that stress-induced OPA1 cleavage has a physiological function, prolonging cell survival by generating S-OPA1 when cells face adverse conditions.
Figure 9.
Silencing OMA1 increases H2O2-induced cell death. A, immunoblots of cell lysate of a stable clone expressing OMA1 shRNA in OPA1-v1 cells. H2O2-induced OPA1 cleavage is attenuated upon OMA1 knockdown (KD) in OPA1-v1 cells. B, quantification of L- and S-OPA1 bands in immunoblotting showing the percentage of the total. C, LDH assays for cell death analyses. Decreasing OPA1 cleavage by OMA1 silencing in OPA1-v1 cells increases H2O2-induced cell death. ***, p = 0.00036; **, p = 0.0012 (Student's t test, unpaired). Error bars, S.E. D, OCRs of OPA1-v1 and OMA1-KD/OPA1-v1 cells in OXPHOS conditions with and without H2O2. Error bars, S.E. E, Cellular respiratory control ratios show significantly lower respiration coupling in OMA1-KD/OPA1-v1 cells treated with H2O2 in OXPHOS conditions, compared with that of OPA1-v1 cells. *, p = 0.0149 (Student's t test, unpaired). Error bars, S.E.
Discussion
OPA1 is associated with the IM, where it performs a dual function, IM fusion and cristae maintenance. OPA1's role for cristae maintenance is critical for the OXPHOS activity of cells, as proper cristae structure is required for functional assembly of respiratory complexes and supercomplexes (5). OPA1 exists in cells as a mixture of transmembrane L-OPA1 and soluble S-OPA1 generated by cleavage of L-OPA1. Mitochondrial and cellular stresses activate OMA1 to induce L-OPA1 cleavage, accumulating S-OPA1. Previous studies on differential functions of L- and S-OPA1 demonstrated that L-OPA1 is competent for mitochondrial fusion, but S-OPA1 is not (5, 12, 13, 24, 25). Regarding OPA1's role for cristae maintenance, different experimental approaches resulted in contradictory information. The studies using the genetic deletions of proteins that control OPA1 cleavage (OMA1, Yme1L, and Phb2) concluded that, similar to the IM fusion function, L-OPA1 is the form that maintains cristae structure, but not S-OPA1 (20, 30). These data created the notion that S-OPA1 is a functionally insignificant cleavage product and thus OPA1 cleavage in stress conditions is causal for mitochondrial fragmentation and energetic deficiency, leading to cell demise (27–29). In contrast, more recent studies using the cell systems that express L- or S-OPA1 exclusively found that S-OPA1 is also capable of maintaining cristae structure and thus energetic competency, indistinguishable from L-OPA1 (5, 25). In the current study, we were able to further distinguish the energetic function of L- and S-OPA1, in which S-OPA1 outperforms L-OPA1 in supporting cell survival in a stress condition.
The functional difference between L- and S-OPA1 was revealed by applying oxidant stress only under OXPHOS condition, where cells expressing L-OPA1 exclusively (and OPA1-KO cells) showed significantly increased sensitivity to H2O2-induced cell death, compared with S-OPA1–containing cells. Adding glucose to the OXPHOS media prevented H2O2-induced cell death in L-OPA1 (OPA1-v1ΔS1) and OPA1-KO cells, indicating that OXPHOS potentiates oxidant injury in these cells. Our data show that OPA1-v1ΔS1 and OPA1-KO cells generate increased levels of superoxide in OXPHOS conditions, which may contribute to sensitizing these cells to H2O2-induced cell death. In addition to superoxide, another contributing factor is the inability of these cells to produce sufficient ATP in OXPHOS conditions. H2O2 causes DNA damage. PARP is a DNA damage sensor and, upon H2O2-mediated DNA damage, is recruited to the DNA lesions, where it rapidly activates poly(ADP)ribosylation of itself and other proteins using NAD+ as a substrate, causing depletion of NAD+, decreased respiration, and ATP production (51–53). Therefore, insufficient ATP in OPA1-KO and L-OPA1–only cells in OXPHOS conditions would result in cellular energy crisis, leading to necrotic cell death. It is interesting that the ATP level of WT cells under H2O2 treatment in the OXPHOS condition is similar to that of OPA1-v1ΔS1 cells. It is likely that increased ROS and oxidative stress in OPA1-v1ΔS1 cells under these conditions also contribute to increased cell death.
Our data indicate that H2O2-induced cell death in OXPHOS conditions is mostly necrotic as judged by the lack of apoptotic cleavage of caspase-3 and PARP-1, in contrast to actD-induced apoptosis. Furthermore, a loss of cytochrome c in OPA1-v1ΔS1 and OPA1-KO cells also indicates necrotic death (Fig. 3B). In support of this notion, H2O2 treatments in glucose-free conditions appear to increase the 74-kDa fragment of PARP-1 (most notable in OPA1-v1), one of the necrotic fragments generated by cathepsins (Fig. 3A) (37, 54).
Our data suggest that there is an intrinsic difference between L- and S-OPA1 in maintaining mitochondrial function, and we found the differential regulation of MPT by L- and S-OPA1. Considering no involvement of ROS in this differential regulation of MPT (Fig. 8, C and D), it is likely that ROS and MPT are independent factors that dictate cell death sensitivity in OXPHOS conditions. These observations also suggest that OPA1 may be a novel regulator of MPT. Based on our data, L- and S-OPA1 may play opposite roles in MPT, in which L-OPA1 increases MPT whereas S-OPA1 renders resistance to MPT. Whereas the normal MPT sensitivity is maintained in WT and OPA1-v1 cells that have both L- and S-OPA1, the lack of L-OPA1 in OPA1-v5 cells decreases the MPT sensitivity, and the lack of S-OPA1 in OPA1-v1ΔS1 cells does the opposite. As for the mechanism of potential OPA1-mediated regulation of MPT, an interaction between OPA1 and complex V (ATP synthase) has been reported (4). The c-ring of complex V as well as the dimeric form of complex V were suggested to function as the PTP (55, 56). It is possible that the OPA1-complex V interaction may regulate MPT. However, gene KO studies have disproved the PTP role of complex V (57, 58). Therefore, the mechanism by which OPA1 may regulate MPT is currently not understood. A previous report suggested that spastic paraplegia 7 (SPG7) is an essential part of the PTP, as the loss of SPG7 greatly increases mCRC, rendering MPT resistance (59). However, SPG7 likely functions as a regulator of PTP by modulating mitochondrial Ca2+ (60, 61). SPG7 is a subunit of IM-associated m-AAA protease that forms heteromeric complex with another m-AAA subunit, AFG3L2 (62). Interestingly, a previous report suggested an involvement of SPG7 in OPA1 cleavage (12). More recently, m-AAA was suggested to regulate OMA1 maturation, contributing to OPA1 cleavage (63). It is possible that SPG7/m-AAA regulates MPT by controlling OPA1 cleavage. Regardless, our data show that L- and S-OPA1 exert opposite effects on MPT sensitivity, which is likely the underlying mechanism by which L- and S-OPA1 cells have different susceptibilities to oxidant insult.
The current study provides new information regarding the functional difference in L- and S-OPA1. We uncovered the differences between L- and S-OPA1 in mitochondrial electron transport activity and in conferring MPT sensitivity. For both, S-OPA1 is protective in oxidant insult, whereas L-OPA1 alone without S-OPA1 renders the opposite effect. Furthermore, we showed that silencing OMA1 in oxidant stress increases cell death, indicating that S-OPA1 generation by OMA1-mediated OPA1 cleavage has a protective role. These new findings challenge the conventional idea that stress-induced OPA1 cleavage is detrimental, causing mitochondrial fragmentation and cell death (26–29). The literature indicates that OMA1 KO causes obesity and energetic defect (64), supporting our findings. In contrast, OMA1-KO mice were protected from heart failure and renal ischemia (26, 65). OMA1 KO can have a pleiotropic effect on multiple cellular processes (66–69); therefore, it would be difficult to interpret whole-organ/animal physio-pathology by OMA1 KO in relation to OPA1 cleavage. Furthermore, our data using OMA1 silencing were obtained using oxidant insults and simple MEF cells that express only one OPA1 variant (OPA1-v1); therefore, it is possible that they may not represent the in vivo effect of OMA1 KO on complex pathological processes of heart failure and renal ischemia.
Regardless of OMA1, our studies using cells exclusively expressing L- or S-OPA1 demonstrated a beneficial role of S-OPA1, not L-OPA1, in cellular stress. However, there are reports that appear to contradict our findings. One study reported that overexpression of a noncleavable form of OPA1 (similar to the v1ΔS1) was protective against retinal ischemia/reperfusion injury (70). However, the conclusion is debatable because, upon insult, there was a greater increase of S-OPA1 in the retina with the L-OPA1 construct used in the study. Another study showed that OMA1/Yme1L double-KO cells that contain only L-OPA1 are resistant to apoptosis induced by 1.0 mm H2O2 (20). Although these data seem to contradict our observation, the study used glucose-containing media in which we did not see much cell death. As discussed above, it also cannot be ruled out that KOs of OMA1 and Yme1L have other effects in addition to deficiency in generating S-OPA1. Regarding OPA1 and MPT, one study showed that cardiac mitochondria from OPA1+/− heterozygous mice had decreased MPT sensitivity to Ca2+ (71). The authors interpreted that this seemingly unexpected result was possibly due to larger mitochondria in the heart or disorganized IM that perturbs IM-OM association that might be necessary for MPT. Whereas our study did not assess the effect of partial decrease of OPA1, we showed that OPA1 KO greatly decreased mCRC, which is consistent with a previous study with OPA1 silencing (72).
Our new notion that S-OPA1 generation is protective can now explain puzzling results from previous studies with mouse retinas of hyperhomocysteinemia (HHcy). HHcy induces excitotoxicity and oxidative stress in retinal ganglion cells (RGCs). HHcy mice showed early death of RGCs, but subsequently the RGC death was attenuated (73), suggesting an activation of a cellular protective mechanism. Intriguingly, despite the occurrence of RGC death in HHcy, the retinal microarray of these mice and additional analyses showed an increase of OPA1 gene expression with a more pronounced increase of S-OPA1 variants (OPA1-v5 and -v8) (73, 74). Gene expression is a costly task, especially for stressed cells. If S-OPA1 is nonfunctional, increasing S-OPA1 expression would be a futile effort for cells under stress. Based on our new findings, it is likely that, in retinal HHcy stress, increased S-OPA1 expression supports RGC survival, attenuating cell death and vision loss. Not only in oxidant insult, our experiments also showed that S-OPA1-containing cells survive better than L-OPA1–only cells in nutrient depletion (results not shown). Therefore, contrary to the conventional notion that OPA1 cleavage occurring in stress conditions is detrimental, our new data suggest that stress-induced OPA1 cleavage is a novel protective mechanism in which the increased S-OPA1 level prolongs cell survival in cells facing adverse conditions.
Experimental procedures
Cell culture
OPA1-KO MEFs were from the American Type Culture Collection (ATCC CRL2995). Stable cell lines expressing each of the human OPA1 variants (OPA1-v1, OPA1-v1ΔS1, or OPA1-v5) in the OPA1-KO MEFs were established by puromycin selection and single-colony isolation as described before (5).
All cell lines were maintained in the complete medium DMEM (Gibco, 11065-065) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin. Puromycin (0.5 μg/ml) was included except for WT and OPA1-KO MEFs. For treatment and survival assays, cells were seeded in the complete medium and then washed three times with Dulbecco's PBS (DPBS) and switched to the test medium: basal DMEM (Sigma–Aldrich, D5030) supplemented with 1 mm pyruvate, 4 mm glutamine, 44 mm sodium bicarbonate, 100 units/ml penicillin, 100 μg/ml streptomycin, and 5% FBS, as glucose-free OXPHOS medium. For glucose-containing medium, glucose (25 mm) was added to glucose-free medium. For oxidant insult, H2O2 was added up to 750 μm to the test medium.
Trypan blue assay
Cells were seeded in a 24-well plate in the complete medium and incubated for 16–24 h. Cells were then washed with DPBS and further incubated in test medium containing 500 μm H2O2 for 18 h. Cells (both floating and adhering) were harvested and stained using trypan blue (Invitrogen). Total and trypan blue–positive cells were counted using an automatic cell counter (Bio-Rad).
Alamar Blue assay
Cells were seeded in a 96-well plate in the complete medium. After overnight incubation, cells were washed with DPBS and further incubated in test medium containing 500 μm H2O2 (90 μl/well) for 16 h. Ten microliters of 10× Alamar Blue stock solution (0.4 mg/ml in PBS) was added to each well, and absorbance was measured at 570 and 600 nm after 1–5 h using a microplate reader (BioTek Instruments). The percentage of reduction of Alamar Blue was calculated by a formula ((117,216 × A570 − 80,586 × A600)/(155,677 × A′600 − 14,652 × A′570) × 100), where 117,216 = molar extinction coefficient of Alamar Blue in the oxidized form at 600 nm; 80,586 = molar extinction coefficient of Alamar Blue in the oxidized form at 570 nm; 14,652 = molar extinction coefficient of Alamar Blue in the reduced form at 600 nm; 155,677 = molar extinction coefficient of Alamar Blue in the reduced form at 570 nm; A600 = absorbance of test wells at 600 nm; A570 = absorbance of test wells at 570 nm; A′600 = absorbance of negative control wells at 600 nm; and A′570 = absorbance of negative control wells at 570 nm (33).
ROS measurements
The level of ROS was detected using the fluorescent probe DHE or MitoSOX (Invitrogen). Cells were loaded with 5 μm DHE or MitoSOX in the test medium without FBS and with or without 25 mm glucose at 37 °C for 30 min. The medium was removed, and fresh test medium containing 5% FBS was added. Images were acquired at room temperature, and mean fluorescence intensity was measured using IPLab imaging software (Scanalytics, Inc.).
LDH-based cytotoxicity assay
Released LDH activity, a surrogate marker of necrosis, was measured using the LDH-Cytotoxicity Assay Kit II (Abcam) according to the manufacturer's instructions. At 30 min at room temperature, final absorbance was measured at 450 nm (reference at 650 nm) using a microplate reader (BioTek Instruments).
GSH assay
Cells grown in complete media were washed and incubated in test media with and without 500 μm H2O2 for 16 h, and total GSH and oxidized GSH (GSSG) was measured using a GSH colorimetric detection kit (Invitrogen) with modifications (75). Cells were washed once with prechilled PBS, scraped in PBS on ice, and pelleted by centrifugation at 1000 × g for 5 min at 4 °C. Pellets were resuspended in ice-cold extraction buffer containing 0.1% Triton X-100 and 0.6% 5-sulfo-salicylic acid, homogenized by sonication, and then frozen at −80 °C. Thawed samples were centrifuged at 3000 × g for 4 min at 4 °C, and supernatant was subjected to determination of total GSH and GSSG according to the manufacturer's protocol. Free glutathione (GSH) concentration was calculated by subtraction of GSSG (1 GSSG = 2 GSH) from total GSH.
Respiration analyses
Oxygen consumption rates (OCRs) were measured using a Seahorse XFe24 Analyzer (Agilent) according to the manufacturer's protocol. Briefly, cells were plated into an XFe24 cell culture microplate in complete media. Cells were then incubated in the test medium with and without 500 μm H2O2 for 14 h and further incubated for 1 h in respiration medium (DMEM with 5 mm pyruvate, 2 mm glutamine, and 5 mm HEPES without glucose and sodium bicarbonate, pH 7.4) at 37 °C in a non-CO2 incubator. For OCR, 2 μm oligomycin, 4 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and 1 μm antimycin A and rotenone were injected in sequence. OCR was normalized by protein amount.
Cellular ATP assay
ATP content was measured using an ATP detection assay kit-Luminescence (Cayman). Cells incubated in the test medium with and without 500 μm H2O2 for 16 h were washed once with prechilled PBS and lysed in ATP detection sample buffer, and ATP concentration was determined according to the manufacturer's instructions. ATP levels were normalized with protein amount.
mCRC analysis
Cells were trypsinized and collected by centrifugation at 156 × g for 3 min and resuspended in respiration buffer (125 mm KCl, 2 mm K2HPO4, 1 mm MgCl2, 20 mm HEPES, pH 7.0) (47). Cells (2 million or more in 0.5 ml) were transferred to the cuvette and permeabilized by adding digitonin to 0.01%. Mitochondria in permeabilized cells were energized by adding 5 mm glutamate and 2.5 mm malate. Thapsigargin (1 μm) was added to inhibit ER Ca2+ uptake. The Ca2+ indicator arsenazo III (100 μm) was added, and kinetic measurements of absorbance were performed at 650 nm (reference at 685 nm) (76–78). After 1 min, 40 μm CaCl2 was added at every 2 min until no further Ca2+ uptake was observed. mCRC was calculated by total Ca2+ amount taken up per million cells.
Cobalt-quenching calcein assay
Cells were loaded with 1.0 μm calcein-acetomethoxy ester (Invitrogen) and 1.5 mm CoCl2 in DPBS for 15 min at room temperature. Mitochondrial calcein fluorescence was verified by MitoTracker Red CMXRos (25 nm, Invitrogen). Calcein fluorescence images were acquired and quantified.
OMA1 silencing
OPA1-v1 cells were infected with lentivirus carrying mouse OMA1 shRNA (proprietary sequence information, sc-151297-V, Santa Cruz Biotechnology, Inc.), and stable cells were obtained by single-colony isolation.
Immunoblot analysis
Protein samples were prepared in Laemmli sample buffer, run on SDS-polyacrylamide gels, and transferred to the polyvinylidene difluoride membrane. The membranes were blocked, incubated with primary antibodies overnight at 4 °C, followed by secondary antibodies, and developed using the chemiluminescence imaging system (Bio-Rad). The following antibodies were used: anti-OPA1 (612606, BD Biosciences; 1:2000), anti-caspase-3 (9662, Cell Signaling; 1:1000), anti-PARP-1 (9542, Cell Signaling; 1:1000), anti-GPx1/2 (sc-133160, Santa Cruz Biotechnology; 1:1000), anti-MnSOD (611580, BD Biosciences; 1:1000), anti-CypD (18466-1-AP, Proteintech; 1:1000), anti-NDUFA5 (GTX111016, GeneTex; 1:500) for complex I, anti-succinate dehydrogenase subunit B (GTX113833, GeneTex; 1:500) for complex II, anti-UQCRC2 (GTX114873, GeneTex; 1:500) for complex III, anti-COX4 (GTX114330, GeneTex; 1:500) for complex IV, anti-ATP synthase subunit β for complex V (A-21351, Thermo Fisher Scientific; 1:500), anti-β-actin (A1978, Sigma; 1:1000), anti-TOM20 (11802-1-AP, Proteintech; 1:1000), anti-OMA1 (sc-515788, Santa Cruz Biotechnology; 1:1000), and anti-α-tubulin (62204, Thermo Scientific; 1:1000). All commercial antibodies have been validated by prior publications.
Statistical analyses
Quantitative data were analyzed by Student's t test for the comparison between two groups (two-tailed, unpaired). For comparison between multiple groups, analysis of variance (ANOVA) was used followed by Tukey's post hoc test. The data are expressed mostly as means ± S.E. unless noted otherwise; p < 0.05 was considered a statistically significant difference.
Data availability
All data are contained within the article.
Author contributions
H. L., S. B. S., and Y. Y. conceptualization; H. L., S. B. S., S.-S. S., and Y. Y. data curation; H. L., S. B. S., and Y. Y. formal analysis; H. L., S. B. S., S.-S. S., and Y. Y. funding acquisition; H. L., S. B. S., and Y. Y. validation; H. L., S. B. S., and Y. Y. investigation; H. L. and S. B. S. methodology; H. L., S. B. S., and Y. Y. writing-original draft; H. L. and Y. Y. project administration; H. L., S. B. S., S.-S. S., and Y. Y. writing-review and editing; S. B. S., S.-S. S., and Y. Y. resources.
Supplementary Material
Acknowledgment
We thank David Chan (California Institute of Technology) for providing OPA1 variant constructs.
This work was supported by American Heart Association Grants 16GRNT31170032 and 19TPA34890037 (to Y. Y.) and National Institutes of Health Grants R01HL093671 (to Y. Y. and S.-S. S.), R01EY028103 and R01EY012830 (to S. B. S.), and R01HL137266 (to S.-S. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S5.
- OXPHOS
- oxidative phosphorylation
- IM
- inner membrane
- OM
- outer membrane
- TM
- transmembrane
- L- and S-OPA1
- long- and short-form OPA1, respectively
- KO
- knockout
- ROS
- reactive oxygen species
- MPT
- mitochondrial permeability transition
- MEF
- mouse embryonic fibroblast
- DMEM
- Dulbecco's modified Eagle's medium
- PARP
- poly(ADP-ribose) polymerase
- actD
- actinomycin D
- DHE
- dihydroethidium
- LDH
- lactate dehydrogenase
- GPx
- GSH peroxidase
- MnSOD
- Mn-superoxide dismutase
- PTP
- permeability transition pore
- CypD
- cyclophilin D
- CsA
- cyclosporine A
- mCRC
- mitochondrial calcium retention capacity
- HHcy
- hyperhomocysteinemia
- RGC
- retinal ganglion cell
- FBS
- fetal bovine serum
- DPBS
- Dulbecco's PBS
- OCR
- oxygen consumption rate
- ANOVA
- analysis of variance.
References
- 1. Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., and Lenaers G. (2003) Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278, 7743–7746 10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- 2. Cipolat S., Martins de Brito O., Dal Zilio B., and Scorrano L. (2004) OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101, 15927–15932 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G. V., Rudka T., Bartoli D., Polishuck R. S., Danial N. N., De Strooper B., and Scorrano L. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- 4. Patten D. A., Wong J., Khacho M., Soubannier V., Mailloux R. J., Pilon-Larose K., MacLaurin J. G., Park D. S., McBride H. M., Trinkle-Mulcahy L., Harper M. E., Germain M., and Slack R. S. (2014) OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 33, 2676–2691 10.15252/embj.201488349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee H., Smith S. B., and Yoon Y. (2017) The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J. Biol. Chem. 292, 7115–7130 10.1074/jbc.M116.762567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee H., and Yoon Y. (2018) Mitochondrial membrane dynamics-functional positioning of OPA1. Antioxidants 7, E186 10.3390/antiox7120186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alexander C., Votruba M., Pesch U. E., Thiselton D. L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., Bhattacharya S. S., and Wissinger B. (2000) OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- 8. Delettre C., Lenaers G., Griffoin J. M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., Astarie-Dequeker C., Lasquellec L., Arnaud B., Ducommun B., Kaplan J., and Hamel C. P. (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- 9. Amati-Bonneau P., Valentino M. L., Reynier P., Gallardo M. E., Bornstein B., Boissière A., Campos Y., Rivera H., de la Aleja J. G., Carroccia R., Iommarini L., Labauge P., Figarella-Branger D., Marcorelles P., Furby A., et al. (2008) OPA1 mutations induce mitochondrial DNA instability and optic atrophy “plus” phenotypes. Brain 131, 338–351 10.1093/brain/awm298 [DOI] [PubMed] [Google Scholar]
- 10. Yu-Wai-Man P., Griffiths P. G., Gorman G. S., Lourenco C. M., Wright A. F., Auer-Grumbach M., Toscano A., Musumeci O., Valentino M. L., Caporali L., Lamperti C., Tallaksen C. M., Duffey P., Miller J., Whittaker R. G., et al. (2010) Multi-system neurological disease is common in patients with OPA1 mutations. Brain 133, 771–786 10.1093/brain/awq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delettre C., Griffoin J. M., Kaplan J., Dollfus H., Lorenz B., Faivre L., Lenaers G., Belenguer P., and Hamel C. P. (2001) Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 109, 584–591 10.1007/s00439-001-0633-y [DOI] [PubMed] [Google Scholar]
- 12. Ishihara N., Fujita Y., Oka T., and Mihara K. (2006) Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song Z., Chen H., Fiket M., Alexander C., and Chan D. C. (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Landes T., Leroy I., Bertholet A., Diot A., Khosrobakhsh F., Daloyau M., Davezac N., Miquel M. C., Courilleau D., Guillou E., Olichon A., Lenaers G., Arnauné-Pelloquin L., Emorine L. J., and Belenguer P. (2010) OPA1 (dys)functions. Semin. Cell Dev. Biol. 21, 593–598 10.1016/j.semcdb.2009.12.012 [DOI] [PubMed] [Google Scholar]
- 15. Olichon A., Emorine L. J., Descoins E., Pelloquin L., Brichese L., Gas N., Guillou E., Delettre C., Valette A., Hamel C. P., Ducommun B., Lenaers G., and Belenguer P. (2002) The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 523, 171–176 10.1016/S0014-5793(02)02985-X [DOI] [PubMed] [Google Scholar]
- 16. Griparic L., Kanazawa T., and van der Bliek A. M. (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178, 757–764 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Head B., Griparic L., Amiri M., Gandre-Babbe S., and van der Bliek A. M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., and Langer T. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M. F., Attardi G., Larsson N. G., Neupert W., and Reichert A. S. (2006) Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281, 37972–37979 10.1074/jbc.M606059200 [DOI] [PubMed] [Google Scholar]
- 20. Anand R., Wai T., Baker M. J., Kladt N., Schauss A. C., Rugarli E., and Langer T. (2014) The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olichon A., Elachouri G., Baricault L., Delettre C., Belenguer P., and Lenaers G. (2007) OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ. 14, 682–692 10.1038/sj.cdd.4402048 [DOI] [PubMed] [Google Scholar]
- 22. Baker M. J., Lampe P. A., Stojanovski D., Korwitz A., Anand R., Tatsuta T., and Langer T. (2014) Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33, 578–593 10.1002/embj.201386474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mishra P., Carelli V., Manfredi G., and Chan D. C. (2014) Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19, 630–641 10.1016/j.cmet.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., Da Cruz S., Clerc P., Raschke I., Merkwirth C., Ehses S., Krause F., Chan D. C., Alexander C., Bauer C., et al. (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Del Dotto V., Mishra P., Vidoni S., Fogazza M., Maresca A., Caporali L., McCaffery J. M., Cappelletti M., Baruffini E., Lenaers G., Chan D., Rugolo M., Carelli V., and Zanna C. (2017) OPA1 isoforms in the hierarchical organization of mitochondrial functions. Cell Rep. 19, 2557–2571 10.1016/j.celrep.2017.05.073 [DOI] [PubMed] [Google Scholar]
- 26. Xiao X., Hu Y., Quirós P. M., Wei Q., López-Otín C., and Dong Z. (2014) OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of ischemic kidney injury. Am. J. Physiol. Renal Physiol. 306, F1318–F1326 10.1152/ajprenal.00036.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wai T., García-Prieto J., Baker M. J., Merkwirth C., Benit P., Rustin P., Rupérez F. J., Barbas C., Ibañez B., and Langer T. (2015) Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science 350, aad0116 10.1126/science.aad0116 [DOI] [PubMed] [Google Scholar]
- 28. Wai T., and Langer T. (2016) Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117 10.1016/j.tem.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 29. MacVicar T., and Langer T. (2016) OPA1 processing in cell death and disease: the long and short of it. J. Cell Sci. 129, 2297–2306 10.1242/jcs.159186 [DOI] [PubMed] [Google Scholar]
- 30. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., and Langer T. (2008) Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 22, 476–488 10.1101/gad.460708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ban T., Heymann J. A., Song Z., Hinshaw J. E., and Chan D. C. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 10.1093/hmg/ddq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Page B., Page M., and Noel C. (1993) A new fluorometric assay for cytotoxicity measurements in-vitro. Int. J. Oncol. 3, 473–476 [PubMed] [Google Scholar]
- 33. Larson E. M., Doughman D. J., Gregerson D. S., and Obritsch W. F. (1997) A new, simple, nonradioactive, nontoxic in vitro assay to monitor corneal endothelial cell viability. Invest. Ophthalmol. Vis. Sci. 38, 1929–1933 [PubMed] [Google Scholar]
- 34. Nicholson D. W., Ali A., Thornberry N. A., Vaillancourt J. P., Ding C. K., Gallant M., Gareau Y., Griffin P. R., Labelle M., and Lazebnik Y. A. (1995) Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376, 37–43 10.1038/376037a0 [DOI] [PubMed] [Google Scholar]
- 35. Tewari M., Quan L. T., O'Rourke K., Desnoyers S., Zeng Z., Beidler D. R., Poirier G. G., Salvesen G. S., and Dixit V. M. (1995) Yama/CPP32 β, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell 81, 801–809 10.1016/0092-8674(95)90541-3 [DOI] [PubMed] [Google Scholar]
- 36. Satoh M. S., and Lindahl T. (1992) Role of poly(ADP-ribose) formation in DNA repair. Nature 356, 356–358 10.1038/356356a0 [DOI] [PubMed] [Google Scholar]
- 37. Chaitanya G. V., Steven A. J., and Babu P. P. (2010) PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 8, 31 10.1186/1478-811X-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Troyano A., Sancho P., Fernández C., de Blas E., Bernardi P., and Aller P. (2003) The selection between apoptosis and necrosis is differentially regulated in hydrogen peroxide-treated and glutathione-depleted human promonocytic cells. Cell Death Differ. 10, 889–898 10.1038/sj.cdd.4401249 [DOI] [PubMed] [Google Scholar]
- 39. Neame S. J., Rubin L. L., and Philpott K. L. (1998) Blocking cytochrome c activity within intact neurons inhibits apoptosis. J. Cell Biol. 142, 1583–1593 10.1083/jcb.142.6.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bobba A., Atlante A., Giannattasio S., Sgaramella G., Calissano P., and Marra E. (1999) Early release and subsequent caspase-mediated degradation of cytochrome c in apoptotic cerebellar granule cells. FEBS Lett. 457, 126–130 10.1016/S0014-5793(99)01018-2 [DOI] [PubMed] [Google Scholar]
- 41. Jemmerson R., LaPlante B., and Treeful A. (2002) Release of intact, monomeric cytochrome c from apoptotic and necrotic cells. Cell Death Differ. 9, 538–548 10.1038/sj.cdd.4400981 [DOI] [PubMed] [Google Scholar]
- 42. Halestrap A. P., Woodfield K. Y., and Connern C. P. (1997) Oxidative stress, thiol reagents, and membrane potential modulate the mitochondrial permeability transition by affecting nucleotide binding to the adenine nucleotide translocase. J. Biol. Chem. 272, 3346–3354 10.1074/jbc.272.6.3346 [DOI] [PubMed] [Google Scholar]
- 43. Vercesi A. E., Kowaltowski A. J., Grijalba M. T., Meinicke A. R., and Castilho R. F. (1997) The role of reactive oxygen species in mitochondrial permeability transition. Biosci. Rep. 17, 43–52 10.1023/A:1027335217774 [DOI] [PubMed] [Google Scholar]
- 44. Baines C. P., Kaiser R. A., Purcell N. H., Blair N. S., Osinska H., Hambleton M. A., Brunskill E. W., Sayen M. R., Gottlieb R. A., Dorn G. W., Robbins J., and Molkentin J. D. (2005) Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature 434, 658–662 10.1038/nature03434 [DOI] [PubMed] [Google Scholar]
- 45. Schinzel A. C., Takeuchi O., Huang Z., Fisher J. K., Zhou Z., Rubens J., Hetz C., Danial N. N., Moskowitz M. A., and Korsmeyer S. J. (2005) Cyclophilin D is a component of mitochondrial permeability transition and mediates neuronal cell death after focal cerebral ischemia. Proc. Natl. Acad. Sci. U.S.A. 102, 12005–12010 10.1073/pnas.0505294102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Basso E., Fante L., Fowlkes J., Petronilli V., Forte M. A., and Bernardi P. (2005) Properties of the permeability transition pore in mitochondria devoid of Cyclophilin D. J. Biol. Chem. 280, 18558–18561 10.1074/jbc.C500089200 [DOI] [PubMed] [Google Scholar]
- 47. Murphy A. N., Bredesen D. E., Cortopassi G., Wang E., and Fiskum G. (1996) Bcl-2 potentiates the maximal calcium uptake capacity of neural cell mitochondria. Proc. Natl. Acad. Sci. U.S.A. 93, 9893–9898 10.1073/pnas.93.18.9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kong D., Xu L., Yu Y., Zhu W., Andrews D. W., Yoon Y., and Kuo T. H. (2005) Regulation of Ca2+-induced permeability transition by Bcl-2 is antagonized by Drp1 and hFis1. Mol. Cell Biochem. 272, 187–199 10.1007/s11010-005-7323-3 [DOI] [PubMed] [Google Scholar]
- 49. Scorrano L., Petronilli V., Di Lisa F., and Bernardi P. (1999) Commitment to apoptosis by GD3 ganglioside depends on opening of the mitochondrial permeability transition pore. J. Biol. Chem. 274, 22581–22585 10.1074/jbc.274.32.22581 [DOI] [PubMed] [Google Scholar]
- 50. Yu T., Sheu S. S., Robotham J. L., and Yoon Y. (2008) Mitochondrial fission mediates high glucose-induced cell death through elevated production of reactive oxygen species. Cardiovasc. Res. 79, 341–351 10.1093/cvr/cvn104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee Y. J., and Shacter E. (1999) Oxidative stress inhibits apoptosis in human lymphoma cells. J. Biol. Chem. 274, 19792–19798 10.1074/jbc.274.28.19792 [DOI] [PubMed] [Google Scholar]
- 52. Filipovic D. M., Meng X., and Reeves W. B. (1999) Inhibition of PARP prevents oxidant-induced necrosis but not apoptosis in LLC-PK1 cells. Am. J. Physiol. 277, F428–E436 10.1152/ajprenal.1999.277.3.F428 [DOI] [PubMed] [Google Scholar]
- 53. Walisser J. A., and Thies R. L. (1999) Poly(ADP-ribose) polymerase inhibition in oxidant-stressed endothelial cells prevents oncosis and permits caspase activation and apoptosis. Exp. Cell Res. 251, 401–413 10.1006/excr.1999.4589 [DOI] [PubMed] [Google Scholar]
- 54. Gobeil S., Boucher C. C., Nadeau D., and Poirier G. G. (2001) Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): implication of lysosomal proteases. Cell Death Differ. 8, 588–594 10.1038/sj.cdd.4400851 [DOI] [PubMed] [Google Scholar]
- 55. Alavian K. N., Beutner G., Lazrove E., Sacchetti S., Park H. A., Licznerski P., Li H., Nabili P., Hockensmith K., Graham M., Porter G. A. Jr., and Jonas E. A. (2014) An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc. Natl. Acad. Sci. U.S.A. 111, 10580–10585 10.1073/pnas.1401591111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giorgio V., von Stockum S., Antoniel M., Fabbro A., Fogolari F., Forte M., Glick G. D., Petronilli V., Zoratti M., Szabó I., Lippe G., and Bernardi P. (2013) Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc. Natl. Acad. Sci. U.S.A. 110, 5887–5892 10.1073/pnas.1217823110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. He J., Carroll J., Ding S., Fearnley I. M., and Walker J. E. (2017) Permeability transition in human mitochondria persists in the absence of peripheral stalk subunits of ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 114, 9086–9091 10.1073/pnas.1711201114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. He J., Ford H. C., Carroll J., Ding S., Fearnley I. M., and Walker J. E. (2017) Persistence of the mitochondrial permeability transition in the absence of subunit c of human ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 114, 3409–3414 10.1073/pnas.1702357114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shanmughapriya S., Rajan S., Hoffman N. E., Higgins A. M., Tomar D., Nemani N., Hines K. J., Smith D. J., Eguchi A., Vallem S., Shaikh F., Cheung M., Leonard N. J., Stolakis R. S., Wolfers M. P., et al. (2015) SPG7 is an essential and conserved component of the mitochondrial permeability transition pore. Mol. Cell 60, 47–62 10.1016/j.molcel.2015.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. König T., Tröder S. E., Bakka K., Korwitz A., Richter-Dennerlein R., Lampe P. A., Patron M., Mühlmeister M., Guerrero-Castillo S., Brandt U., Decker T., Lauria I., Paggio A., Rizzuto R., Rugarli E. I., et al. (2016) The m-AAA protease associated with neurodegeneration limits MCU activity in mitochondria. Mol. Cell 64, 148–162 10.1016/j.molcel.2016.08.020 [DOI] [PubMed] [Google Scholar]
- 61. Hurst S., Baggett A., Csordas G., and Sheu S. S. (2019) SPG7 targets the m-AAA protease complex to process MCU for uniporter assembly, Ca2+ influx, and regulation of mitochondrial permeability transition pore opening. J. Biol. Chem. 294, 10807–10818 10.1074/jbc.RA118.006443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Koppen M., Metodiev M. D., Casari G., Rugarli E. I., and Langer T. (2007) Variable and tissue-specific subunit composition of mitochondrial m-AAA protease complexes linked to hereditary spastic paraplegia. Mol. Cell Biol. 27, 758–767 10.1128/MCB.01470-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Consolato F., Maltecca F., Tulli S., Sambri I., and Casari G. (2018) m-AAA and i-AAA complexes coordinate to regulate OMA1, the stress-activated supervisor of mitochondrial dynamics. J. Cell Sci. 131, jcs213546 10.1242/jcs.213546 [DOI] [PubMed] [Google Scholar]
- 64. Quirós P. M., Ramsay A. J., Sala D., Fernández-Vizarra E., Rodríguez F., Peinado J. R., Fernández-Garcia M. S., Vega J. A., Enríquez J. A., Zorzano A., and López-Otin C. (2012) Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31, 2117–2133 10.1038/emboj.2012.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Acin-Perez R., Lechuga-Vieco A. V., Del Mar Muñoz M., Nieto-Arellano R., Torroja C., Sánchez-Cabo F., Jiménez C., González-Guerra A., Carrascoso I., Benincá C., Quiros P. M., López-Otin C., Castellano J. M., Ruíz-Cabello J., Jiménez-Borreguero L. J., and Enríquez J. A. (2018) Ablation of the stress protease OMA1 protects against heart failure in mice. Sci. Transl. Med. 10, eaan4935 10.1126/scitranslmed.aan4935 [DOI] [PubMed] [Google Scholar]
- 66. Desmurs M., Foti M., Raemy E., Vaz F. M., Martinou J. C., Bairoch A., and Lane L. (2015) C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Mol. Cell Biol. 35, 1139–1156 10.1128/MCB.01047-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bohovych I., Fernandez M. R., Rahn J. J., Stackley K. D., Bestman J. E., Anandhan A., Franco R., Claypool S. M., Lewis R. E., Chan S. S., and Khalimonchuk O. (2015) Metalloprotease OMA1 fine-tunes mitochondrial bioenergetic function and respiratory supercomplex stability. Sci. Rep. 5, 13989 10.1038/srep13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kaser M., Kambacheld M., Kisters-Woike B., and Langer T. (2003) Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 278, 46414–46423 10.1074/jbc.M305584200 [DOI] [PubMed] [Google Scholar]
- 69. Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., and Winge D. R. (2012) Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287, 7289–7300 10.1074/jbc.M111.313148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sun Y., Xue W., Song Z., Huang K., and Zheng L. (2016) Restoration of Opa1-long isoform inhibits retinal injury-induced neurodegeneration. J. Mol. Med. (Berl.) 94, 335–346 10.1007/s00109-015-1359-y [DOI] [PubMed] [Google Scholar]
- 71. Piquereau J., Caffin F., Novotova M., Prola A., Garnier A., Mateo P., Fortin D., Huynh le H, Nicolas V., Alavi M. V., Brenner C., Ventura-Clapier R., Veksler V., and Joubert F. (2012) Down-regulation of OPA1 alters mouse mitochondrial morphology, PTP function, and cardiac adaptation to pressure overload. Cardiovasc. Res. 94, 408–417 10.1093/cvr/cvs117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kushnareva Y. E., Gerencser A. A., Bossy B., Ju W. K., White A. D., Waggoner J., Ellisman M. H., Perkins G., and Bossy-Wetzel E. (2013) Loss of OPA1 disturbs cellular calcium homeostasis and sensitizes for excitotoxicity. Cell Death Differ. 20, 353–365 10.1038/cdd.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ganapathy P. S., Moister B., Roon P., Mysona B. A., Duplantier J., Dun Y., Moister T. K., Farley M. J., Prasad P. D., Liu K., and Smith S. B. (2009) Endogenous elevation of homocysteine induces retinal neuron death in the cystathionine-β-synthase mutant mouse. Invest. Ophthalmol. Vis. Sci. 50, 4460–4470 10.1167/iovs.09-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ganapathy P. S., Perry R. L., Tawfik A., Smith R. M., Perry E., Roon P., Bozard B. R., Ha Y., and Smith S. B. (2011) Homocysteine-mediated modulation of mitochondrial dynamics in retinal ganglion cells. Invest. Ophthalmol. Vis. Sci. 52, 5551–5558 10.1167/iovs.11-7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rahman I., Kode A., and Biswas S. K. (2006) Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 1, 3159–3165 10.1038/nprot.2006.378 [DOI] [PubMed] [Google Scholar]
- 76. Brookes P. S., Salinas E. P., Darley-Usmar K., Eiserich J. P., Freeman B. A., Darley-Usmar V. M., and Anderson P. G. (2000) Concentration-dependent effects of nitric oxide on mitochondrial permeability transition and cytochrome c release. J. Biol. Chem. 275, 20474–20479 10.1074/jbc.M001077200 [DOI] [PubMed] [Google Scholar]
- 77. Tokunaga H., Hollenberg N. K., and Graves S. W. (2000) Sodium-dependent calcium release from vascular smooth muscle mitochondria. Hypertens. Res. 23, 39–45 10.1291/hypres.23.39 [DOI] [PubMed] [Google Scholar]
- 78. Nadtochiy S. M., Burwell L. S., and Brookes P. S. (2007) Cardioprotection and mitochondrial S-nitrosation: effects of S-nitroso-2-mercaptopropionyl glycine (SNO-MPG) in cardiac ischemia-reperfusion injury. J. Mol. Cell Cardiol. 42, 812–825 10.1016/j.yjmcc.2007.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article.